Rep:Mod:01330404
Ammonia
General information
Ammonia NH3
calculation method: RB3LYP
basis set:6-31G(d.p)
final energy E in a.u.:-56.55776873
RMS Gradient Norm:0.00000485
point group:C3V
optimized NH3 bond length B=1.01798 (N-H)
optimized NH3 bond angle A=105.741 (H-N-H)
3D Jmol model
test molecule |
Link to the .log file
File:YUEHE PHUNT NH3 OPTF POP.LOG
Item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986275D-10
Optimization completed.
-- Stationary point found.
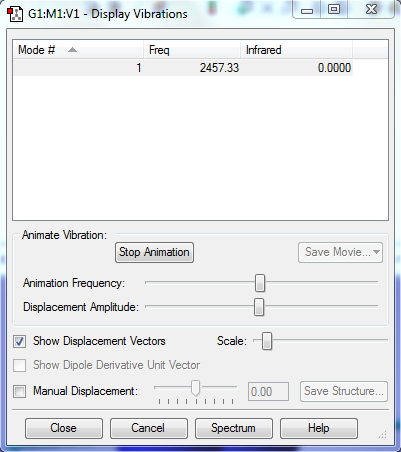
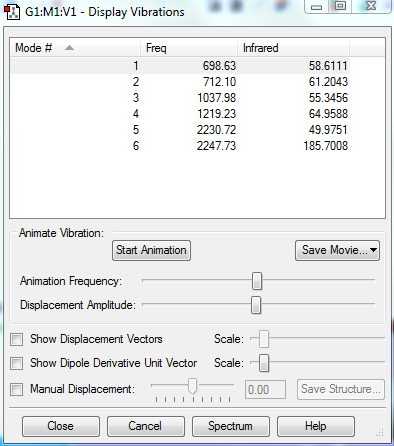
Vibration table
Modes and interpretation
Number of modes from 3N-6 rule:3*4-6=6 modes
Degenerate modes: mode2&3, mode5&6
Bending: mode1,2&3
Stretching: mode4,5&6
Highly symmetric mode:4
Umbrella mode:1
bands expected:4
Charge on N:-1.125
Charge on H:0.375
Negative charge on N is expected as N is more electronegative.
Positive charge on H is expected.
Nitrogen
General information
Nitrogen N2
calculation method: RB3LYP
basis set:6-31G(d.p)
final energy E in a.u.:-109.52359111
RMS Gradient Norm:0.02473091
point group:D*H
3D Jmol Model
test molecule |
Link to the .log file
Item table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401079D-13
Optimization completed.
-- Stationary point found.
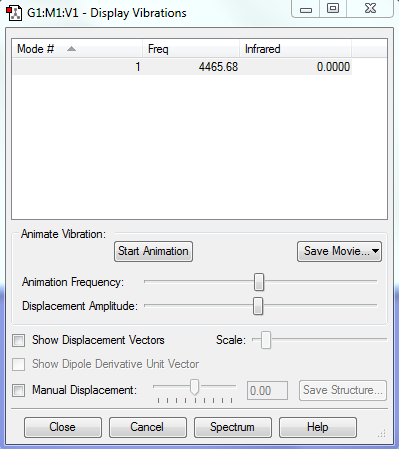
Vibration table
Hydrogen
General information
Hydrogen H2
calculation method: RB3LYP
basis set:6-31G(d.p)
final energy E in a.u.:-1.15928020
RMS Gradient Norm:0.09719500
point group:D*H
3D Jmol model
test molecule |
Link to the .log file
Item table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
Vibration table
Energy calculation
E(NH3)=-56.55776873
2*E(NH3)=-113.1155375
E(N2)=:-109.52359111
E(H2)=:-1.15928020
3*E(H2)=-3.4778406
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.11410579=-299.58 kJ/mol
As ΔE is negative the ammonia product is more stable.
Molecule selected: F2
General information
Fluorine F2
calculation method: RB3LYP
basis set:6-31G(d.p)
final energy E in a.u.:-199.42620785
RMS Gradient Norm:0.23253407
point group:D*H
Bond length: 1.16000
Charge on F atoms: 0.000
3D Jmol Model
test molecule |
Link to the .log file
Item table
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995025D-08
Optimization completed.
-- Stationary point found.
Vibration table
MO images and descriptions of F2





Figure.1:Sigma bonding MO formed by two 2s orbitals. It is relatively deep in energy and is occupied by two electrons. It enhances the bonding character of the bond.
Figure.2:Sigma anti-bonding MO formed by two 2s orbitals; Higher energy than the previous bonding MO. It is occupied by two electrons and therefore enhances the anti-bonding character of the bond.
Figure.3:Pi bonding MO formed by two 2p orbitals. It is occupied by two electrons and is still negative in energy. There is no pi bonds in F2 as the effect of pi bonding MOs is cancelled by the occupied pi anti-bonding MOs.
Figure.4:HOMO of F2 molecule formed by two 2p orbitals. It is a pi anti-bonding MO and is occupied by two electrons.
Figure.5:LUMO of F2 molecule formed by two 2p orbitals. It is a sigma anti-bonding MO and is unoccupied of electrons.
Independence calculation:H2SiO
General information
H2SiO
calculation method: RB3LYP
basis set:6-31G(d.p)
final energy E in a.u.:-365.89319642
RMS Gradient Norm:0.03065697
point group:CS
Charge on Si atom: 1.472
Charge on H atom(s): -0.236
Charge on O atom: -1.001
3D Jmol Model
test molecule |
Link to the .log file
Item table
Item Value Threshold Converged?
Maximum Force 0.000023 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000023 0.001800 YES
RMS Displacement 0.000017 0.001200 YES
Predicted change in Energy=-5.109741D-10
Optimization completed.
-- Stationary point found.