Rep:Mod2:hr610
Module 2: Bonding and Molecular Orbitals in Main Group Compounds
Introduction
Chemistry is known worldwide as an experimental subject. The constant decreasing cost and increasing accuracy and power of computing software has stimulated chemists to explore the area of computational chemistry. [1] It has recently been reported that computational methods were used in the design of cheaper catalysts for the removal of ethyne from streams of ethylene gas. [2]
In this project we will use the program; Gaussian to optimise structures, carry out frequency analysis, determine vibrational modes, predict the Molecular Orbtials (MO's) and estimate the charge distribution on several molecules. The compounds that we will study in this module are commonly used in industry, such as boranes in numerous laboratory syntheses and thallium bromide in semiconductors.
- Accuracy
All errors stated below are assumed, therefore in the following exercises;
- Energy will have an error of ≈ 10 kJ/mol ~ 3.8 x10-3 a.u/ Hartree, which are the units that will be reported by the program Gaussview.
An important point is that the total energy of a particular molecule cannot be compared to other molecules, and these will be reported in a.u. Instead, Relative energies can be used to compare molecules, and will be reported in kJ/mol. Molecules can only be compared using their relative energies when they both have the same number and type of atoms. Also, they must have been computed using the same method/basis set.
- Dipole moments will be accurate to 2 decimal places ~ 0.01 Deby.
- Vibrational frequencies will be accurate to 0 decimal places and have a systematic error of about 10%. This is because a harmonic approximation is used for anharmonic vibrations in the frequency calculations carried out. Therefore, the error in vibrational frequencies will vary in magnitude, depending on the size of the peak under study.
- Intensities are given to the nearest whole number, but the accuracy is much less than this.
- Bond distances are accurate to ~ 0.01Å.
- Bond angles are accurate to ≈ 0.1°.
Optimisation of a Molecule of BH3
We will be using the computational program Gaussian to compute the optimisation. In carrying out an optimization we are determining the optimium position of the nuclei for a given electronic configuration- the first step for any quantum chemical calculation. The primary method used for most of the calculations in this module is based on density functional theory (DFT), specifically B3LYP. The basis set for this optimisation will be 3-21G. We begin by setting the geometry of BH3 to trigonal planar with a bond length of 1.50A.
The optimisation will be done by adopting the Bohn-Oppenheimer approximation, assuming a set position for the nuclei and solving the Schrödinger equation for the electron density and the electrons. The force is the first derivative of the potential energy so at a set position there is a given potential energy with a corresponding force. This force is applied to the molecule in steps until an energy minima is reached which represents the geometry on a potential energy diagram where the total force sums to zero. Thus, the optimised geometry is obtained and can be used to compute physical properties of the molecule, such as the optimum distance of the hydrogen nuclei from the boron centre in this configuration. The overall borane molecule adopts the D3H point group.

| ||
|---|---|---|
As apparent in the computed borane molecules, the models look identical before and after the optimisation. However, the B-H bond distances have decreased after the optimisation. In comparison to literature values reported by Schuurman et al:[3] and other literature [4] the BH3 optimised structure shows an exact match for the B-H bond lengths and angles.
The summary table shows that the calculation type is FOPT, the calculation method used was RB3LYP, the basis set is 3-21G, the final energy in atomic units is -26.46 a.u., the dipole moment is 0.00 Debye and the gradient is 0.00 a.u. The near zero gradient (the exact number presented is 0.00004507 a.u.) shows the optimisation has successfully completed as the energy minimum has been found.
The output data from the LOG file (follow link below) confirms all the parameters have converged successfully. Thus, the forces have converged: F = − V / r and the atom coordinates were optimised so that a small displacement will not result in a noticeable energy change. Hence, the molecule is at its optimum geometry- the one present in the gas-phase, with the dihedral angles between the B-H bonds at 120 degrees -the optimum angle for a planar molecule. Also, the optimum bond length for the B-H bonds is 1.19 Å.
File:BH3 OPTIMISATION3-21g.LOG

The optimisation was captured by iterative steps that modelled the intermediate geometries, and they are shown below in Fig.1
| Stage 1 | Stage 2 | Stage 3 | Stage 4 |
|---|---|---|---|

|

|
|
|
| 1.5Å | 1.41Å | 1.23Å | 1.19Å |

In the first two structures, the molecule appears to have no bonds. Gaussview only includes bonds that are shorter than some pre-determined length. The molecule is being optimised from a length of 1.5Å to 1.19Å. Thus, Gaussview only includes bonds in the final two structures. The definition of a bond is not based on distance but on electron density. One can say; "A bond is a region of high electron density between two atoms leading to an electromagnetic force of attraction between the atoms."
The graphs (See images on the right) reveal that Gaussview carried out four stages of optimisation until a stationary point was found. The Total Energy curve, shows Gaussview traversing the potential energy surface (PES) of BH3 and finding the structure that corresponds to the energy minima in the graph. The second graph is the RMS (Root Mean Square or average) Gradient, and shows the gradient going to zero as we approach a turning point (maximum or minimum) in the total energy curve. To conclude that the turning point is a minimum, the sign of the second derivative of the energy curve needs to be calculated using analysis of the vibrational frequencies. This analysis is carried out below.[5]
A Better basis-set
A basis set is a set of mathematical functions, normally representing atomic orbitals (AOs). At first, these AOs were used in the Hartree-Fock method and were represented as Slater orbitals, which corresponded to a set of functions which decayed exponentially with distance from the nuclei. Later, Frank Boys approximated the Slater orbitals as a linear combination of Gaussian orbitals that are much easier to compute, as they reduce the 4-centre integrals found in Slater orbitals to finite sums of 2-centre integrals and then 1-centre integrals. These are around 5 orders of magnitude quicker to solve than Slater orbitals, so Gaussian basis functions are much cheaper and easier to use. [6] [7]
There are numerous different basis sets that can be used, here we will use 'Split Valence Basis Sets:'
Split Valence Basis Sets can be used for complex systems - they increase the number of basis functions per atom. In this module we have used the 3-21G above and now will use the more accurate 6-31G set. By increasing the number of primitives devoted to the core and first valence function, the 6-31G basis set improves upon 3-21G energetics at the expense of increased computer times. [7]

| |
|---|---|
The Summary table shows the gradient at near zero and so the optimisation is concluded to have gone to completion.
However, as seen in this summary table- the point group is reported as CS. After repeating this optimisation 3 times, each time restricting the point group to D3H- the same 'CS' result was reported. However, I can confirm that the point group is D3H as the B-H bond lengths were all identical and the H-B-H bond angles were all 120°. Therefore, this error is likely to be due to the program software; Gaussview.

Again the output file (follow link below) confirms that all the parameters have successfully converged. File:BH3 OPT BASIS6-31G.LOG
| 3-21G Total energy (a.u) | 6-31G Total energy (a.u) |
| -26.46226433 | -26.61532264 |
The table shows that when using the 6-31G basis set, the total energy is lower (more negative) than that reported from the 3-21G basis set. The energy difference of 0.15305831 a.u (about 402 KJ/mol) is large and this does not mean that the 6-31G(d,p) computation is a better method. The total energy for any calculation is highly dependent on the quality of the basis set. Thus, we can never compare the energy of structures computed using different basis sets (or pseudo-potentials).
Part 1.1: Frequency analysis Of BH3
A frequency calculation must use the same model and basis set that produced the optimised geometry. For this analysis, DFT was used again with the B3LYP functional and 6-31G basis set for accuracy.
 | |||
|---|---|---|---|
| |||
See link below for the frequency LOG output file: File:HR610 BH3 FREQ.LOG


The total energy of the system was found to be the same as the energy recorded for the Optimisation. This confirmed the Optimisation had been performed on the correct file. As apparent in the Log file for the frequency analysis- the frequencies are all positive and so an energy minimum is concluded to be found and therefore the optimum geometry of the molecule. The image to the right showing the 'Low frequencies' represent the motion of the center of mass (the central atom). This atom is the heaviest and so it does not vibrate much and thus the frequencies are low.
As reported in the table, the symmetry of each vibration was determined using a representation table.
The lack of any negative stretching frequencies shows the optimised structure is in fact an energy minimum. In addition to this, the Optimised structure makes chemical sense - one would expect BH3 to adopt a Trigonal planar structure.
- IR Spectrum of BH3

Only 3 stretches are observed in the spectrum of BH3, whereas 6 vibrational modes have been computed. This can be explained as two sets of four E' Stretching modes are degenerate (1212cm-1 and 2722cm-1), and thus appear at the same frequency on the IR spectrum. Also, the A1' stretching mode does not appear on the spectrum, as no change in dipole is caused by this stretch and the requirement for a mode to appear on an IR spectrum is for a change in dipole moment to occur. The three peaks on the spectrum are; a strong one at 1161cm-1 (wagging), a weak one at 1212cm-1(rocking) and another strong one at 2722cm-1 (degenerate asymmetric stretch).
The correlation with the literature values that state the peaks to be at 1206, 1387 and 1995 cm-1 suggest that the predicted vibrational frequencies are not perfect but are within a reasonable degree of accuracy. [8] This could have been improved by using a better basis set for the optimisation.
Part 1.2: Population Analysis: MO Diagram Of BH3
The energy of the Optimised structure of BH3 was calculated using DFT-B3LYP, a 6-31G basis set and full NBO and population analysis. 20467
A MO diagram was created in Chemdraw as shown below. The fragment orbitals of H3 were constructed using a representation table, the reduction and projection formula. Hydrogen and Boron are of similar electropositivity and therefore of similar energy. The fragment orbitals of H3 all have bonding character, so the a1' is placed below the 1s orbital of Boron. The e' levels of H3 are of comparable energy to the boron p orbitals. Then, orbitals of the same symmetry can interact and mix together. Additionally, secondary orbital mixing can occur between molecular orbitals of the same symmetry, but this is insignificant for BH3 as there are no orbitals of the same symmetry that are close in energy.[9]
The MO's from the population analysis were visualised in Gaussview and added to the following diagram.
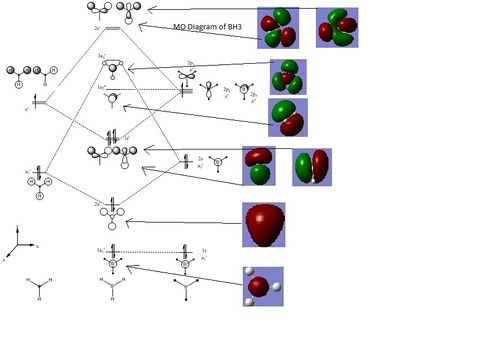
The phase distribution in both the real and LCAO MOs are very similar, suggesting that both theories work well in predicting relative energies for simple molecules. This similarity is harder to see for the MO's of higher energies as these orbitals are more diffused.
It is difficult to predict how stabilised/destabilised the bonding and anti-bonding orbitals are respectively, but computational chemistry can determine the energies of these orbitals relative to one another.
A limitation of qualitative MO theory is apparent if we consider the 3a1’ orbital. This orbital shows no differences in size between the various orbital contributions. For anti-bonding orbitals we expect to see less electron density on the more electronegative atom (hydrogen) and therefore a smaller orbital. However this is not visible in the calculated MO.
Optimising a Molecule of TlBr3
A useful approximation is to only consider the valence electrons which dominate in bonding interactions. For this reason all core electrons that are non-bonding can be grouped together into an overall effect that is modelled by a function called a pseudo-potential. Pseudo potentials are special functions that model the core electrons of an atom, thereby simplifying the calculation and allowing us to focus only on the valence electrons. The LACVP series of basis sets is a combination of the 6-31G basis set with the LANL2DZ pseudopotential. The atoms H to Ar in the periodic table can be described with the 6-31G basis set while the heavier atoms (such as Tl) are modeled using the LANL2DZ basis set.[10].[11]
"The pseudopotential approximation is certainly the most successful of all approximate relativistic molecular theories.” Werner Kutzelnigg, 1987[12]
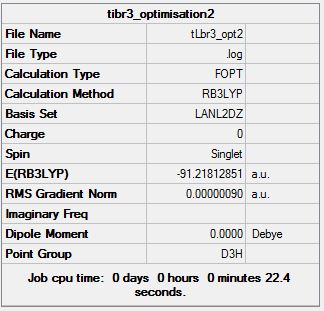
TlBr3 has more electrons (81/Tl and 35/Br) than the molecule previously modelled: BH3. Also, the atoms are much heavier and so pseudo potentials will be used for this computation. Again the method of choice will be the DFT- B3LYP. We start from the Trigonal planar structure with restricted D3h symmetry. We restrict the symmetry to ensure a good quality of results and no “splitting” of frequencies, thus reducing any errors and allowing any further vibrational analysis to be credible and accurate.
Results after optimisation:
Link to D-Space for TLBr3 Optimisation:
DOI 20451
 | |||
|---|---|---|---|
| |||
The Tl-Br bond length obtained after this optimisation (2.65 Å ± 0.005) can be concluded to be reasonably accurate in comparison to literature values.[13] However, this could have been further improved by using a more accurate basis set. The bond angles of Trigonal planar complexes and their dipole moments are expected to be 120° and 0 D, respectively and this is what was reported from the Optimisation.

The results to the right show that the optimisation was successfully carried out as all the parameters have converged. However, as for BH3, a confirmation that a minimum has been reached is needed by carrying out a frequency analysis.
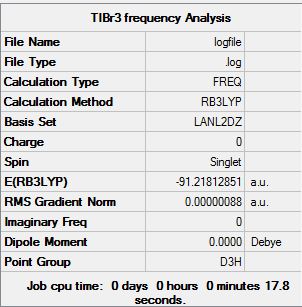
Frequency analysis of TlBr3
Frequency analysis was performed on the optimised structure of TlBr3 using the same method and basis set.
D-Space link to TlBr3 frequency analysis:
DOI 20458
 | |||
|---|---|---|---|
| |||

The image to the right shows that all the vibrations of TlBr3 were found to be positive. The absence of negative frequencies concludes that a minimum has been reached.
Hence, the structure of TlBr3 has therefore been successfully Optimised using pseudo potentials.
IR Spectrum of TlBr3
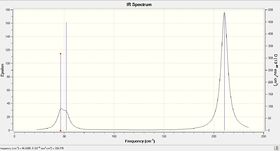
Vibrational values of TlBr3:
Comparison of Vibrations of BH3 and TlBr3
Since TlBR3 and BH3 have the same point group, their IR active stretching modes of vibration will be the same. What differs is the magnitude of the frequency. This may be explained by the fact that Thallium is much heavier which results in slower vibrations and thus shorter frequencies. The totally symmetric (dipole=0D) stretch at 165.27cm-1 has an intensity of zero as expected due to its lack of dipole moment and is not observed. The degenerate bends at 46.43cm-1 overlap with the bend at 52.14cm-1 reducing the number of peaks observed. The other two degenerate vibrations at 210.69cm-1 correspond to asymmetric stretches which are naturally higher in energy than the bends.
Optimisation of a molecule of BBr3
When compounds contain both heavy atoms which require a pseudo-potential, and light atoms, which are treated more accurately with a full basis set we need to be able to mix psuedo-potentials and basis sets. Therefore, we will select the method ‘GEN’ to allow us to specify the basis sets for each individual atom. In addition to this we add "pseudo=cards gfinput" to the calculation to allow us to also specify pseudo-potentials for each atom individually. This was done by editing the input file and setting the heavier Br atoms to pseudo-potentials and the lighter B atom to the basis set 6-31G for accuracy.[14]
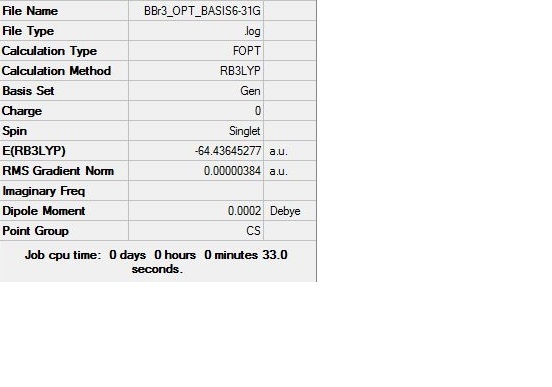 | |||
|---|---|---|---|
| |||
Log file for the 6-31G optimisation: File:BBR3 OPT BASIS6-31G.LOG

The Optimised B-Br bond length is in good comparison to the literature value that states the value to be 1.87 +/- 0.02Å. Also, the Br-B-Br bond angle of 120° is in exact agreement to the literature value. [15] Thus, the optimisation is further concluded to have been carried out accurately.
;Summary of bond lengths:
| BH3 (Å) | BBr3(Å) | TlBr3(Å) | |
| 1.19 | 1.93 | 2.65 |
From this table we can see that from substituting the Hydrogen ligands to Bromine the bond length increases quite significantly. This is because hydrogen only consists of s-type orbitals resulting in a good overlap with the Boron. Also, the larger difference in electronegativity between Boron and Bromine, compared to Boron and Hydrogen, will give rise to weaker bonds as orbitals that are different in energy have less stabalisation when combined than those closer in energy. In addition to this, Bromine is a larger atom than hydrogen with more diffused orbitals and a poorer overlap with Boron- thus increasing the bond length.
Also, from substituting the Boron atom for Thallium the bond length increases further to 2.65Å. Thallium and Boron are both in group 13 of the periodic table, but Thallium is a larger atom with a larger covalent radius and therefore more diffused orbitals with poorer overlaps and so this will result in an increase in the bond length.
Analysis of NH3
NBO Analysis:
Natural bond orbital (NBO) analysis has proven to be an invaluable tool, facilitating the interpretation of electronic structure calculations in a chemically intuitive manner. NBO analysis replaces the delocalised canonical molecular orbitals with a set of transferable localized bonding orbitals and lone pairs, with a strong connection to the ubiquitous Lewis structures of general chemistry. The analysis is “ natural” since it is based solely on the density matrix, obtained from the underlying electronic structure calculation. This lack of artificial parameters helps ensure robust, physically motivated results and chemical insights into bonding and reactivity.[16]
The NBO method encompasses a suite of algorithms that enable fundamental bonding concepts to be extracted from Hartree-Fock or Density Functional Theory computations. NBO analyses of selected molecules that span the periodic table illustrate the deciphering of the molecular wavefunction in terms commonly understood by chemists: Lewis structures, charge, bond order, bond type, hybridization, resonance, donor–acceptor interactions.[17]
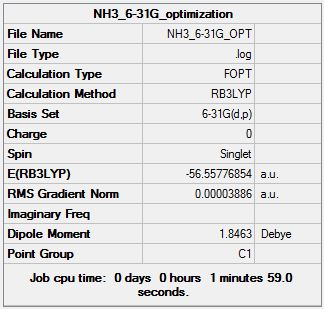
The NBO method investigates the charge distribution and the molecular orbitals in a more localised manner. Firstly, we optimised the NH3 molecule using the 6-31G basis set.
Log File: File:NH3 6-31G OPT.LOG
 | |||
|---|---|---|---|
| |||
Again, the summary file shows a near zero gradient and the log file concludes that the structure was successfully Optimised as the parameters had all converged.

As for previous molecules; a frequency analysis was carried out to show that the frequencies were all positive and thus the energy minimum was obtained.
Frequency analysis Log File: File:NH3 6-31G FREQ.LOG

- Population Analysis
Link to D-Space for NH3 Population Analysis: 20597
- Results of the NBO Analysis
Gaussview generated a charge distrbution picture of the molecule, which is displayed below to show the spread of electron density around NH3.
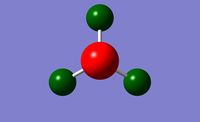
|
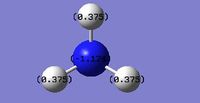
|
|||
| NBO Charge Distribution for NH3 | Specific NBO charges for NH3 |
The brighter the red colour, the more negative the charge and the brighter the green colour, the more positive the charge. The nitrogen atom clearly represents the localisation of negative charge which is what is expected for this electronegative atom. The charge distribution is also in good agreement with the Pauling electronegativity scale, as Nitrogen is more electronegative than Hydrogen (N = 3.04, H = 2.10). The Charge Distribution range was set from -1.000 to 1.000.
The output log file used to produce the MOs contains relevant and useful information about the natural bond orbitals (NBOs). The NBO analysis partitions the electron density of the whole molecule out into atomic like orbitals, which are then used to form 2e-2c bonds.
- Bond Coefficient Data

- The first three natural bond orbitals (BD) are between nitrogen and hydrogen. 69% of the bond is contributed from the N orbitals which have a hybridisation of 25%s+75%p, while 31% of the bond comes from the H orbital which is 100%s.
- The 4th Natural Bond orbital is a core orbital centered on Nitorgen, which is 100% s character. This will be the occupied 1s orbital of Nitrogen, which does not participate in bonding.
- Thus the N has formed 4 sp3 hybrid orbitals which each interact with the sAO of one hydrogen atom.
- The last orbital shown (Orbital 5) is the nitrogen lone pair (LP) 25% s abd 75% p orbital. This is formally occupied so it has an occupation close to 2.00.
Energy analysis of Ammonia-Borane.
Hydrogen (H2) has been proposed as an alternative power source, both as a transportation fuel and, through its use in a PEM fuel cell, as a replacement for batteries for portable electricity. Ammonia− Borane (NH3BH3) has garnered interest as a hydrogen storage material due to its high weight percent hydrogen content and ease of H2 release relative to metal hydrides. [18] In this module we will compute the dative bond energy between the Nitrogen and Boron atoms. A point to consider is when computing energies the reactants (NH3 and BH3) and products (NH3BH3) must be computed at exactly the same level, this includes basis set and method.
A molecule of NH3BH3 was optimised at the B3LYP/6-31G(d,p) level: File:NH3BH3 OPT6-31G.LOG
 | |||
|---|---|---|---|
|
To conclude the stationary point found is a minimum; a frequency analysis was carried out:
Frequency Analysis of NH3BH3:
This frequency analysis showed no negative frequencies and therefore confirmed that we had an energy minimum.
| Units | BH3 Total energy | NH3 Total energy | NH3BH3 Total energy | Energy Difference | |
| AU | -26.61532264 | -56.55776854 | -83.22469032 | 0.05159914 | |
| KJ/mol | 135.4735421 |
The conversion factor used was 1AU = 2625.5 KJ/mol
To conclude: The bond dissociation energy of NH3BH3 was found to be 135.47KJ/mol. In comparison to literature values that state the value to be 130KJ/mol we can conclude that the optimisations of the three molecules were accurately carried out and that the final bond energy obtained is highly accurate. [19]
Mini Project:
Optimisation of N(CH3)4+,P(CH3)4+, S(CH3)3+,N(CH3)3(CH2OH)+ and N(CH3)3(CH2CN)+
- Links to D-Space for Optimisations:
N(CH3)4+: 20630
P(CH3)4+: 20632
S(Ch3)3 : 20634
N(CH3)3(CH2OH)+ 20670
N(CH3)3(CH2CN)+ 20677

|

|

|

|

|
||||||
| Summary table for N(CH3)4+ | Summary table for P(CH3)4+ | Summary table for S(CH3)3+ | Summary table for N(CH3)3(CH2OH)+ | Summary table for N(CH3)3(CH2CN)+ |
|
|
|
|
|
|||||||||||||||||||||
| N(CH3)4+ | P(CH3)4+ | S(CH3)3+ | N(CH3)3(CH2OH)+ | N(CH3)3(CH2CN)+ |
- The geometries of N(CH3)4+, P(CH3)4+ and S(CH3)3+ show some similarity's and differences as the central molecule is changed.
- N(CH3)4+ and P(CH3)4+ both have the same C-N-C and C-P-C bond angle of 109.5° which is the expected bond angle for a tetrahedral geometry.
- S(CH3)3+ has a central C-S-C bond angle of 102.7° which suggests that it has a distorted trigonal pyramidal structure due to its lone pair. Also, it only has 3 CH3 groups. This is because sulphur has more electrons than nitrogen and phosphorus and therefore to maintain the single positive charge- only three CH3 groups are present.
- The bond lengths of the C-X bond reveal that C-N bond is the shortest and therefore strongest bond and this is because nirtogen has the least diffused orbitals and therefore the best orbital overlap with carbon that is in the same group as Nitrogen.
| N(CH3)4+ | P(CH3)4+ | S(CH3)3+ | N(CH3)3(CH2OH)+ | N(CH3)3(CH2CN)+ |
Also, the relevant section of the output files shown above show that all the parameters converged and that the Optimisation was successful. To confirm that the turning point corresponds to a minimum value; a frequency analysis was next run on each structure.
Frequency Analysis
D-Space links for FREQUENCY ANALYSIS:
[N(CH3)4]+: 20636
[P(CH3)4]+: 20637
[S(CH3)3]+: 20639
N(CH3)3(CH2OH)+ 20682
N(CH3)3(CH2CN)+
20685
Low frequencies --- -15.2335 -6.4001 -0.0002 0.0005 0.0010 7.4852 Low frequencies --- 179.0378 278.2890 285.2908 |
N(CH3)4+ |
Low frequencies --- -19.4387 -8.5108 -0.0017 0.0028 0.0032 11.9452 Low frequencies --- 151.4829 181.6655 189.5408 |
P(CH3)4+ |
Low frequencies --- -10.8446 -8.1192 0.0020 0.0022 0.0025 23.3223 Low frequencies --- 158.3466 193.9565 198.7377 |
S(CH3)3+ |
Low frequencies --- -6.7614 0.0008 0.0010 0.0011 13.1719 16.0343 Low frequencies --- 130.6057 211.1266 255.8290 |
N(CH3)3(CH2OH)+ |
Low frequencies --- -4.2325 0.0004 0.0006 0.0007 6.2084 14.0549 Low frequencies --- 91.1074 153.8165 210.7687 |
N(CH3)3(CH2CN)+ |
The frequency analysis concluded that the Optimised structure was a minimum for all five molecules. This is known as the frequency values are all positive.
Population Analysis of N(CH3)4+,P(CH3)4+ and S(CH3)3+
The next step of the module was to carry out a population Analysis of all three molecules.
D-Space links for Population Analysis:
N(CH3)4 MO : 20653
P(CH3)4 MO : 20652
S(CH3)3 MO : 20654
The molecular orbitals of N(CH3)4 were visualized in gaussview and a selection are presented below.
NBO Analysis of the molecules

|

|
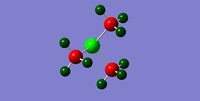
|
||||
| NBO Charge Distribution for N(CH3)4+ | NBO Charge Distribution for P(CH3)4+ | NBO Charge Distribution for S(CH3)3+ |
The NBO analysis carried out on these three molecules is expected to help rationalize the change in charge density throughout the molecule. The green regions indicate high positive charges while the red regions indicate high negative charges. The more negative the charge the greater the electron density about the region. The patterns observed are explained below.
NBO Specific Charges:

|

|

|
||||
| NBO Specific Charge Distribution for N(CH3)4+ | NBO Specific Charge Distribution for P(CH3)4+ | NBO Specific Charge Distribution for S(CH3)3+ |
The Pauling Electronegativity values for the atoms involved are: N: 3.0 P: 2.19 S: 2.58 C: 2.55 H: 2.10
This information can be used to help explain the charge distribution observed in the cations:
- The N(CH3)4+ molecule shows the Nitrogen atom to have a charge of -0.295 and the carbon atoms to have charges of -0.483. This would suggest that the carbons have more electron density than the nitrogen atoms which is in disagreement with the Pauling electronegativity values above. This can be explained by the compound having a positive charge, making the nitrogen more electropositive than expected. The Hydrogen atoms have a charge of 0.269 which is the most electropositive charge in the molecule- this agrees with the fact that hydrogen is the most electropositive atom within the molecule.
- The P(CH3)4+ shows the Phosphorus atom to have a charge of 1.667, the carbons -1.06 and the hydrogen atoms 0.298. The most electronegative atom in this molecule is the carbon atom which can explain the observation that the carbon atoms are the areas of the highest electron density. The phosphorus atom is slightly more electronegative than hydrogen and so we would expect the hydrogen's to have a more positive charge density than phosphorus. However, this is not observed as the compound has a positive charge of +1 that is centralized on the phosphorus atom, thus decreasing its electron density.
- The S(CH3)3+ shows the sulfur atom to have a charge of 0.917, carbons -0.845 and the hydrogen atoms 0.297. Sulfur is the most electronegative atom in this compound but has a similar value to carbons (difference of only 0.03) and so the large difference in the charge distribution values can be explained by the positive charge that is localized on the sulfur atom and therefore making this atom more electropositive that would be expected from Pauling values alone. The charge on the sulfur is not exactly +1 as it is slightly more EN than carbon and so witdraws some electron density from the carbon atoms.
- The hydrogen atoms in all 3 molecules show extremely similar charge values from 0.269 to 0.298 and therefore changing the central atom is shown to have no effect on the electron density of the hydrogen atoms.
- The electron density on the central atoms increases in the order of P<S<N which follows the order of the Pauling Electronegativity values with Nitrogen being the most electronegative central atom and therefore having the most electron density out of the three compounds.
- The carbon atoms within the molecules show increasing electron density in the order of N(CH3)4+<S(CH3)3+<P(CH3)4+. This also can be explained by the electronegativities of the central atom. Nitrogen is the most EN and so withdraws more electron density from the adjacent carbon atoms than P or S. Phosphorus is the least EN atom and so withdraws less electron density from the adjacent carbon atoms resulting these carbons having the highest electron density.
Using the NBO population analysis compare and contrast the relative contribution of the C and heteroatom to the C-X bond. How do your results relate to the charge distribution just studied?
The relevant sections of the population analyses are displayed below.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99118) BD ( 1) C 1 - H 2
( 63.47%) 0.7967* C 1 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.1154
0.0004 -0.6891 0.0206 0.4965 0.0165
-0.0044 0.0025 -0.0171 -0.0140 -0.0032
( 36.53%) 0.6044* H 2 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0033 0.0156 -0.0166
2. (1.99118) BD ( 1) C 1 - H 3
( 63.47%) 0.7967* C 1 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.6778
-0.0095 0.5234 -0.0008 0.0362 0.0246
0.0187 -0.0016 -0.0014 0.0057 -0.0117
( 36.53%) 0.6044* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0172 -0.0144 -0.0052
3. (1.99118) BD ( 1) C 1 - H 4
( 63.47%) 0.7967* C 1 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.7209
0.0152 0.4138 0.0011 0.2091 0.0216
-0.0159 -0.0055 0.0026 0.0107 -0.0109
( 36.53%) 0.6044* H 4 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0174 -0.0117 -0.0095
4. (1.98451) BD ( 1) C 1 - N 17
( 33.65%) 0.5801* C 1 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4551 -0.0237 0.0026 -0.0818
-0.0035 -0.2806 -0.0119 -0.8389 -0.0356
0.0020 0.0061 0.0210 -0.0032 0.0340
( 66.35%) 0.8146* N 17 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 0.0796
0.0000 0.2733 -0.0001 0.8177 -0.0001
0.0009 0.0027 0.0092 -0.0014 0.0149
5. (1.99118) BD ( 1) C 5 - H 6
( 63.47%) 0.7967* C 5 s( 26.41%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5139 0.0032 -0.0004 -0.6974
-0.0087 0.4387 -0.0242 0.2362 -0.0061
-0.0178 -0.0087 0.0066 0.0044 -0.0082
( 36.53%) 0.6044* H 6 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0209 -0.0081 -0.0055
6. (1.99118) BD ( 1) C 5 - H 7
( 63.47%) 0.7967* C 5 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5140 -0.0032 0.0004 0.0586
0.0200 0.2568 0.0119 0.8156 -0.0125
0.0005 0.0003 -0.0094 0.0013 -0.0208
( 36.53%) 0.6044* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 -0.0050 -0.0092 -0.0205
7. (1.99118) BD ( 1) C 5 - H 8
( 63.47%) 0.7967* C 5 s( 26.41%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5139 -0.0032 0.0004 -0.1385
0.0235 0.6645 0.0047 -0.5234 0.0112
0.0074 -0.0060 0.0183 0.0096 -0.0026
( 36.53%) 0.6044* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 -0.0002 -0.0193 0.0126
8. (1.98452) BD ( 1) C 5 - N 17
( 33.65%) 0.5801* C 5 s( 20.78%)p 3.80( 79.05%)d 0.01( 0.16%)
-0.0003 -0.4553 0.0237 -0.0026 -0.6982
-0.0297 -0.5455 -0.0232 -0.0634 -0.0027
-0.0339 -0.0039 -0.0031 -0.0085 0.0200
( 66.35%) 0.8145* N 17 s( 25.00%)p 3.00( 74.96%)d 0.00( 0.03%)
0.0000 -0.5000 0.0007 0.0000 0.6805
-0.0001 0.5318 -0.0001 0.0617 0.0000
-0.0149 -0.0017 -0.0013 -0.0037 0.0088
9. (1.99117) BD ( 1) C 9 - H 10
( 63.47%) 0.7967* C 9 s( 26.41%)p 2.78( 73.54%)d 0.00( 0.05%)
0.0000 0.5139 0.0032 -0.0004 0.6842
-0.0157 0.5104 0.0120 0.0778 -0.0175
0.0175 0.0049 0.0036 0.0083 -0.0106
( 36.53%) 0.6044* H 10 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0163 -0.0162 0.0008
10. (1.99119) BD ( 1) C 9 - H 11
( 63.47%) 0.7967* C 9 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5140 -0.0032 0.0004 0.0761
0.0023 0.2938 -0.0262 0.8015 0.0019
-0.0014 -0.0027 -0.0156 0.0032 -0.0162
( 36.53%) 0.6044* H 11 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 -0.0025 -0.0037 -0.0226
11. (1.99117) BD ( 1) C 9 - H 12
( 63.47%) 0.7967* C 9 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.7145
0.0090 0.4015 0.0139 0.2506 -0.0206
-0.0132 -0.0119 0.0061 0.0110 -0.0070
( 36.53%) 0.6044* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0183 -0.0135 -0.0035
12. (1.98452) BD ( 1) C 9 - N 17
( 33.65%) 0.5801* C 9 s( 20.78%)p 3.80( 79.05%)d 0.01( 0.16%)
0.0003 0.4552 -0.0237 0.0026 0.1205
0.0051 -0.6988 -0.0297 0.5350 0.0227
-0.0075 0.0057 -0.0333 -0.0211 0.0018
( 66.35%) 0.8145* N 17 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 -0.1175
0.0000 0.6809 -0.0001 -0.5217 0.0001
-0.0033 0.0025 -0.0146 -0.0093 0.0008
13. (1.99119) BD ( 1) C 13 - H 14
( 63.47%) 0.7967* C 13 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5140 -0.0032 0.0004 -0.0956
-0.0205 0.6674 0.0013 -0.5292 0.0166
0.0006 -0.0011 0.0196 0.0113 -0.0037
( 36.53%) 0.6044* H 14 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 0.0061 -0.0188 0.0119
14. (1.99119) BD ( 1) C 13 - H 15
( 63.47%) 0.7967* C 13 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5140 -0.0032 0.0004 0.1015
-0.0240 0.2606 0.0084 0.8102 -0.0071
-0.0025 -0.0077 -0.0096 0.0008 -0.0191
( 36.53%) 0.6044* H 15 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 0.0013 -0.0087 -0.0213
15. (1.99118) BD ( 1) C 13 - H 16
( 63.47%) 0.7967* C 13 s( 26.41%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5139 0.0032 -0.0004 0.6580
0.0106 0.5449 -0.0227 0.0687 -0.0085
0.0198 0.0032 0.0028 0.0000 -0.0107
( 36.53%) 0.6044* H 16 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0201 -0.0113 -0.0005
16. (1.98451) BD ( 1) C 13 - N 17
( 33.65%) 0.5801* C 13 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4552 -0.0237 0.0026 -0.7374
-0.0313 0.4333 0.0184 0.2401 0.0102
-0.0285 -0.0158 0.0093 0.0159 -0.0158
( 66.35%) 0.8146* N 17 s( 24.99%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.4999 -0.0007 0.0000 0.7186
-0.0001 -0.4225 0.0001 -0.2343 0.0000
-0.0125 -0.0069 0.0041 0.0070 -0.0070
|
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98386) BD ( 1) C 1 - H 2
( 64.78%) 0.8048* C 1 s( 24.88%)p 3.02( 75.07%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0070 0.0005 0.8380
0.0075 0.0148 0.0184 0.2188 -0.0093
-0.0040 0.0113 -0.0027 0.0150 -0.0075
( 35.22%) 0.5935* H 2 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0214 -0.0031 -0.0039
2. (1.98391) BD ( 1) C 1 - H 3
( 64.78%) 0.8049* C 1 s( 24.89%)p 3.02( 75.06%)d 0.00( 0.04%)
0.0001 -0.4989 0.0071 -0.0005 0.0719
-0.0099 0.0125 -0.0185 0.8630 0.0064
-0.0021 -0.0057 -0.0055 0.0007 -0.0190
( 35.22%) 0.5935* H 3 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 -0.0003 0.0024 -0.0219
3. (1.98389) BD ( 1) C 1 - H 4
( 64.78%) 0.8049* C 1 s( 24.89%)p 3.02( 75.07%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0071 0.0005 -0.3912
0.0108 0.7073 0.0166 0.3112 -0.0095
-0.0134 -0.0099 0.0108 -0.0039 -0.0046
( 35.22%) 0.5935* H 4 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 0.0079 -0.0196 -0.0061
4. (1.98027) BD ( 1) C 1 - P 17
( 59.56%) 0.7717* C 1 s( 25.21%)p 2.96( 74.70%)d 0.00( 0.08%)
0.0002 0.5018 0.0172 -0.0020 -0.3727
0.0068 -0.7058 0.0130 0.3313 -0.0061
0.0177 -0.0083 -0.0158 -0.0121 -0.0081
( 40.44%) 0.6359* P 17 s( 24.98%)p 2.97( 74.17%)d 0.03( 0.85%)
0.0000 0.0001 0.4998 -0.0007 0.0000
0.0000 0.3709 -0.0006 0.0000 0.7034
-0.0011 0.0000 -0.3308 0.0005 0.0562
-0.0264 -0.0501 -0.0385 -0.0257
5. (1.98384) BD ( 1) C 5 - H 6
( 64.79%) 0.8049* C 5 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4987 0.0070 -0.0005 0.0742
0.0217 0.0129 0.0004 0.8629 0.0034
0.0000 0.0031 -0.0005 -0.0015 -0.0204
( 35.21%) 0.5934* H 6 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 -0.0050 -0.0004 -0.0214
6. (1.98382) BD ( 1) C 5 - H 7
( 64.79%) 0.8049* C 5 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 -0.3738
-0.0209 -0.7076 0.0015 0.3314 -0.0066
0.0083 -0.0037 -0.0143 -0.0109 -0.0048
( 35.21%) 0.5934* H 7 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 0.0122 0.0169 -0.0071
7. (1.98381) BD ( 1) C 5 - H 8
( 64.79%) 0.8049* C 5 s( 24.87%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 -0.3933
-0.0208 0.7059 -0.0023 0.3120 -0.0066
-0.0090 -0.0038 0.0137 -0.0108 -0.0055
( 35.21%) 0.5934* H 8 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 0.0126 -0.0168 -0.0066
8. (1.98032) BD ( 1) C 5 - P 17
( 59.57%) 0.7718* C 5 s( 25.25%)p 2.96( 74.67%)d 0.00( 0.08%)
0.0002 0.5022 0.0171 -0.0020 0.8359
-0.0153 0.0145 -0.0003 0.2181 -0.0040
0.0008 0.0123 0.0002 0.0235 -0.0117
( 40.43%) 0.6358* P 17 s( 25.02%)p 2.96( 74.12%)d 0.03( 0.85%)
0.0000 0.0001 0.5002 -0.0010 0.0000
0.0000 -0.8328 0.0011 0.0000 -0.0147
0.0000 0.0000 -0.2179 0.0003 0.0027
0.0391 0.0007 0.0748 -0.0373
9. (1.98387) BD ( 1) C 9 - H 10
( 64.78%) 0.8049* C 9 s( 24.88%)p 3.02( 75.07%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0071 0.0005 -0.3724
0.0029 -0.7068 0.0023 0.3346 0.0216
0.0149 -0.0034 -0.0066 -0.0096 -0.0077
( 35.22%) 0.5934* H 10 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 0.0086 0.0168 -0.0113
10. (1.98383) BD ( 1) C 9 - H 11
( 64.78%) 0.8049* C 9 s( 24.87%)p 3.02( 75.09%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 0.8378
-0.0003 0.0149 0.0003 0.2195 0.0220
0.0006 0.0028 0.0001 0.0185 -0.0089
( 35.22%) 0.5935* H 11 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0203 -0.0004 -0.0086
11. (1.98386) BD ( 1) C 9 - H 12
( 64.78%) 0.8049* C 9 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0071 0.0005 -0.3917
0.0030 0.7067 -0.0016 0.3122 0.0217
-0.0155 -0.0031 0.0058 -0.0090 -0.0080
( 35.22%) 0.5934* H 12 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 0.0091 -0.0169 -0.0108
12. (1.98030) BD ( 1) C 9 - P 17
( 59.58%) 0.7719* C 9 s( 25.25%)p 2.96( 74.67%)d 0.00( 0.08%)
-0.0002 -0.5022 -0.0171 0.0020 0.0731
-0.0013 0.0146 -0.0003 0.8608 -0.0158
-0.0001 -0.0042 -0.0008 -0.0002 -0.0287
( 40.42%) 0.6358* P 17 s( 24.99%)p 2.97( 74.16%)d 0.03( 0.85%)
0.0000 -0.0001 -0.4999 0.0008 0.0000
0.0000 -0.0721 0.0001 0.0000 -0.0143
0.0000 0.0000 -0.8580 0.0012 -0.0002
-0.0134 -0.0026 -0.0005 -0.0913
13. (1.98385) BD ( 1) C 13 - H 14
( 64.79%) 0.8049* C 13 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4988 0.0071 -0.0005 0.0718
-0.0105 0.0161 0.0184 0.8631 0.0058
0.0020 -0.0059 0.0043 0.0005 -0.0193
( 35.21%) 0.5934* H 14 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 -0.0002 -0.0031 -0.0218
14. (1.98381) BD ( 1) C 13 - H 15
( 64.78%) 0.8049* C 13 s( 24.87%)p 3.02( 75.09%)d 0.00( 0.04%)
-0.0001 0.4986 -0.0070 0.0005 0.8381
0.0080 0.0152 -0.0185 0.2186 -0.0087
0.0051 0.0111 0.0031 0.0146 -0.0076
( 35.22%) 0.5934* H 15 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0215 0.0024 -0.0040
15. (1.98384) BD ( 1) C 13 - H 16
( 64.78%) 0.8049* C 13 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0071 0.0005 -0.3712
0.0113 -0.7084 -0.0165 0.3328 -0.0091
0.0129 -0.0100 -0.0114 -0.0043 -0.0040
( 35.22%) 0.5934* H 16 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 0.0073 0.0196 -0.0067
16. (1.98032) BD ( 1) C 13 - P 17
( 59.58%) 0.7719* C 13 s( 25.25%)p 2.96( 74.67%)d 0.00( 0.08%)
0.0002 0.5022 0.0171 -0.0020 -0.3923
0.0072 0.7047 -0.0129 0.3097 -0.0057
-0.0186 -0.0082 0.0147 -0.0116 -0.0089
( 40.42%) 0.6358* P 17 s( 25.01%)p 2.96( 74.14%)d 0.03( 0.85%)
0.0000 0.0001 0.5001 -0.0009 0.0000
0.0000 0.3903 -0.0006 0.0000 -0.7025
0.0010 0.0000 -0.3092 0.0004 -0.0592
-0.0260 0.0468 -0.0368 -0.0283
|
1. (1.98720) BD ( 1) C 1 - H 2
( 64.83%) 0.8052* C 1 s( 26.49%)p 2.77( 73.46%)d 0.00( 0.05%)
0.0000 -0.5147 0.0056 -0.0006 0.3902
-0.0116 0.7125 0.0088 0.2728 -0.0080
-0.0146 -0.0099 -0.0113 0.0037 0.0059
( 35.17%) 0.5931* H 2 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0011 -0.0071 -0.0211 -0.0058
2. (1.98719) BD ( 1) C 1 - H 3
( 64.83%) 0.8052* C 1 s( 26.49%)p 2.77( 73.46%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 0.8116
0.0027 0.0320 -0.0143 -0.2730 0.0080
0.0059 -0.0145 -0.0039 0.0139 -0.0059
( 35.17%) 0.5931* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 -0.0220 0.0030 0.0058
3. (1.99411) BD ( 1) C 1 - H 4
( 64.23%) 0.8014* C 1 s( 27.23%)p 2.67( 72.72%)d 0.00( 0.05%)
0.0001 0.5219 -0.0018 0.0010 -0.0494
0.0107 0.0800 -0.0172 0.8473 0.0075
-0.0021 -0.0047 0.0076 -0.0010 0.0193
( 35.77%) 0.5981* H 4 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 -0.0010 0.0016 -0.0226
4. (1.98634) BD ( 1) C 1 - S 13
( 48.68%) 0.6977* C 1 s( 19.73%)p 4.06( 80.14%)d 0.01( 0.14%)
0.0003 0.4439 0.0140 -0.0033 -0.4307
0.0033 0.6952 -0.0054 -0.3638 -0.0098
-0.0238 0.0127 -0.0204 -0.0118 -0.0096
( 51.32%) 0.7164* S 13 s( 16.97%)p 4.86( 82.40%)d 0.04( 0.63%)
0.0000 0.0001 0.4118 -0.0076 0.0012
0.0000 0.4274 -0.0188 0.0000 -0.6900
0.0305 0.0000 0.4041 0.0259 -0.0443
0.0327 -0.0527 -0.0220 -0.0051
5. (1.98728) BD ( 1) C 5 - H 6
( 64.82%) 0.8051* C 5 s( 26.52%)p 2.77( 73.43%)d 0.00( 0.05%)
0.0000 0.5149 -0.0057 0.0006 -0.4219
-0.0134 0.6936 -0.0056 -0.2738 0.0079
-0.0106 0.0048 -0.0142 -0.0108 -0.0058
( 35.18%) 0.5931* H 6 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 0.0147 -0.0167 0.0058
6. (1.98727) BD ( 1) C 5 - H 7
( 64.82%) 0.8051* C 5 s( 26.51%)p 2.77( 73.44%)d 0.00( 0.05%)
0.0000 -0.5149 0.0057 -0.0006 0.3770
0.0138 0.7195 -0.0047 0.2726 -0.0079
-0.0091 -0.0039 -0.0145 0.0121 0.0059
( 35.18%) 0.5931* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0011 -0.0136 -0.0176 -0.0058
7. (1.99413) BD ( 1) C 5 - H 8
( 64.22%) 0.8014* C 5 s( 27.25%)p 2.67( 72.70%)d 0.00( 0.05%)
0.0001 0.5220 -0.0018 0.0010 0.0939
-0.0202 0.0037 -0.0006 0.8472 0.0076
0.0002 0.0089 0.0003 0.0023 0.0193
( 35.78%) 0.5982* H 8 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 0.0018 0.0000 -0.0226
8. (1.98627) BD ( 1) C 5 - S 13
( 48.64%) 0.6974* C 5 s( 19.66%)p 4.08( 80.20%)d 0.01( 0.14%)
0.0003 0.4432 0.0141 -0.0033 0.8180
-0.0066 0.0257 -0.0002 -0.3635 -0.0097
0.0017 -0.0240 -0.0007 0.0265 -0.0096
( 51.36%) 0.7167* S 13 s( 16.91%)p 4.88( 82.46%)d 0.04( 0.63%)
0.0000 0.0001 0.4112 -0.0074 0.0012
0.0000 -0.8118 0.0355 0.0000 -0.0254
0.0011 0.0000 0.4036 0.0260 0.0031
-0.0619 -0.0019 0.0493 -0.0051
9. (1.98719) BD ( 1) C 9 - H 10
( 64.83%) 0.8052* C 9 s( 26.49%)p 2.77( 73.46%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 0.8122
0.0018 0.0185 0.0145 -0.2726 0.0079
-0.0041 -0.0147 0.0029 0.0145 -0.0059
( 35.17%) 0.5931* H 10 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 -0.0218 -0.0044 0.0058
10. (1.98720) BD ( 1) C 9 - H 11
( 64.83%) 0.8052* C 9 s( 26.50%)p 2.77( 73.46%)d 0.00( 0.05%)
0.0000 -0.5147 0.0056 -0.0006 0.4337
-0.0110 -0.6867 -0.0095 0.2733 -0.0080
0.0150 -0.0106 0.0107 0.0019 0.0058
( 35.17%) 0.5931* H 11 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0011 -0.0084 0.0206 -0.0058
11. (1.99411) BD ( 1) C 9 - H 12
( 64.23%) 0.8014* C 9 s( 27.24%)p 2.67( 72.72%)d 0.00( 0.05%)
0.0001 0.5219 -0.0018 0.0010 -0.0448
0.0096 -0.0826 0.0178 0.8473 0.0075
0.0019 -0.0042 -0.0078 -0.0013 0.0193
( 35.77%) 0.5981* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 -0.0009 -0.0016 -0.0226
12. (1.98633) BD ( 1) C 9 - S 13
( 48.68%) 0.6977* C 9 s( 19.72%)p 4.06( 80.14%)d 0.01( 0.14%)
-0.0003 -0.4438 -0.0140 0.0033 0.3864
-0.0030 0.7209 -0.0056 0.3638 0.0098
-0.0221 -0.0114 -0.0212 0.0147 0.0096
( 51.32%) 0.7164* S 13 s( 16.96%)p 4.86( 82.41%)d 0.04( 0.63%)
0.0000 -0.0001 -0.4118 0.0076 -0.0012
0.0000 -0.3833 0.0169 0.0000 -0.7155
0.0315 0.0000 -0.4041 -0.0259 -0.0412
-0.0293 -0.0547 0.0274 0.0051
|
||||
| N(CH3)4+ | P(CH3)4+ | S(CH3)3+ |
From analysis of the above files the following summary can be presented that shows the relative contributions of the central atoms to the C-X bonds:
66% N and 34% C for C-N bonds 40% P and 60% C for C-P bonds 61% S and 49%C for C-S bonds
- These results show that the central heteroatom (X) contributes most to the C-X bond in increasing order of P< S< N. This mirrors the order of Pauling electronegativity values and explains the charge distributions observed above.
- The C-X bond has the least electron density on the C atom in the nitrogen molecule molecules because the C-N bond has the highest contribution from the N atom and so the electron density is likely to be concentrated on the N atom rather than the C atom (explained by Nitrogens high electronegativity).
- This is the opposite effect for the C-P bond that has the highest contribution from the C atom instead of the phosphorus atom and so the electron density is likely to be concentrated on the C atom rather than the phosphorus atom. This is reinforced in the above analysis with the fact that the central hetereoatom with the most electron density is the nitrogen and the C atoms of the molecules with the most electron density is the Carbons in the phosphorus molecules.
[NR4]+ (R=alkyl) is often depicted as shown below, with the positive charge placed on the nitrogen center. Based on your results for [N(CH3)4]+, how valid is this description?
This description is not accurate because the NBO analysis shows the Nitrogen atom to have a charge of -0.295 and therefore the positive charge of +1 is not placed directly on the nitrogen. From analysis of the charge distribution throughout the molecule, the positive charge is located on the hydrogen atoms within the molecule which is common for a quaternary ammonium salt.
;Population analysis of [N(CH3)3(ch2OH)]+ and [N(CH3)3(ch2CN)]+
[N(CH3)3(ch2OH)]+ Population Analysis : D-Space link for Population Analysis: 20690
[N(CH3)3(ch2CN)]+ Population Analysis : D-Space link for Population Analysis: 20692
Qu) OH is an electron donating group and CN is an electron withdrawing group, what effect have these groups had on the charge distribution?
- Carbon and nitrogen atoms are meta directors and OH is a para ortho director. CN is purely an electron withdrawing group whereas OH can donate electrons through its lone pair and it is also capable of withdrawing electrons through sigma bonds.
- Oxygen has a very high electron density as shown by the very negative charge from the NBO analysis (-0.725). This is because oxygen is a highly electronegative atom and so withdraws electron density from the H atom of the OH group through the sigma bond. This also explains the low electron density of this hydrogen as it has the most positive charge out of all the atoms of the molecule. In addition to this the C adjacent to the OH group has a lower electron density compared to the other carbons in the molecule and thus the OH group also withdraws electrons from this carbon atom through the sigma bond.
- The OH group also has a lone pair and so can act as an electron donating group and this is observed in the NBO analysis charge distribution whereby the central Nitrogen atom has a charge of -0.322 which has more electron density than the nitrogen in the CN complex.
- The central nitrogen in the CN complex has less electron density as CN is an electron withdrawing group and so withdraws electrons from the central nitrogen atom and the CH2 adjacent to the CN group.
Qu) Compare and contrast the HOMO and LUMO of [N(CH3)4]+, [N(CH3)3(CH2OH)]+ and [N(CH3)3(CH2CN)]+ how has the shape of the orbitals changed? Has the energy of the orbitals moved? What chemical impact could these changes have?
HOMO and LUMO energy differences:
- The OH HOMO is highest in energy as its electron donation effect through its lone pairs dominates and results in higher energy HOMO orbital. CN containing molecule has a higher HOMO than the N(CH3)4+ cation because it has more electrons and therefore filled MO's than the N(Ch3)4+ cation.
- The energy for the LUMO's of the three molecules changes. The N and OH containing molecules have LUMO's very close in energy whereas CN has the LUMO quite a bit lower in energy. This can be explained through the fact that the two compounds with the CN and OH groups having more electrons and therefore more filled molecular orbitals- suggesting higher energies for these frontier orbitals. However, this suggestion does not explain the fact that the N(ch3)4+ complex has a LUMO higher in energy than the CN containing complex. This can however be explained through the fact that CN is an electron withdrawing group and therefore lowering the energy of the molecular orbitals as the electrons are held more tightly and are harder to remove.
- The chemical effect from these different energy values is that it will be easier to remove an electron from the HOMO of [N(Ch3)3(Ch2OH)]+ than the other two complexes and it will also be easier to donate an electron into the LUMO of the [N(Ch3)3(Ch2OH)]+ as this LUMO has the highest energy value.
Shapes of the molecular orbitals:
- The N(Ch3)4+ HOMO has electron density throughout the molecule and so an electron can be expected to be removed from different atomic orbitals of the molecule.
- The CN containing molecule HOMO has electron density concentrated on the CN pi bond and therefore if an electron was to be removed from thsi molecule it would be from this bond.
- The OH containing molecule HOMO has electron density concentrated on the OH group which is likely to be due to the lone pair of electrons on the oxygen atom.
- The LUMO's of the three molecules show similar electron density patterns throughout the molecule.
Conclusion
During this computational assignment many new concepts were explored and introduced, the main one being the use of the program; GaussView to set up and run calculations.
To conclude; good reliable results were obtained throughout the module and were backed up by referenced literature values.
References
- ↑ Grant, G. H.; Richards, W. G.; Computational Chemistry, Oxford Science Publications, 1995, pp 1-4
- ↑ J. Nørskov et al., Chemistry World, 2008 link: http://www.rsc.org/chemistryworld/News/2008/June/05060802.asp
- ↑ M.S. Schuurman, W.D. Allen, H.F. Schaefer III, J. Comput. Chem., 2005, 26, 1106: DOI:10.1002/jcc.20238
- ↑ Fourier transform infrared spectroscopy of the BH, v3 band,Kentarou Kawaguchi, J. Chem. Phys., Vol. 96, No. 5,1 March 1992
- ↑ http://www.huntresearchgroup.org.uk/teaching/teaching_comp_lab_year3/3b_understand_opt.html
- ↑ Young D.C. ‘'Computational Chemistry: A practical guide for applying techniques to real-world problems’', John Wiley & Sons, Inc; 2001, 336
- ↑ 7.0 7.1 Davidson, Ernest; Feller, David (1986). "Basis set selection for molecular calculations". Chem. Rev. 86 (4): 681–696. doi:10.1021/cr00074a002
- ↑ The Raman Spectra of Boron Trifluoride, Trichloride, and Tribromide. The Effect of the Boron Isotopes, J. Chem. Phys. 4, 703 (1936); doi:10.1063/1.1749772
- ↑ Year 2 MO course notes, Dr Patricia Hunt
- ↑ ^ Schwerdtfeger, P. (August 2011), "The Pseudopotential Approximation in Electronic Structure Theory", ChemPhysChem, doi:10.1002/cphc.201100387
- ↑ Grant, G. H.; Richards, W. G.; Computational Chemistry, Oxford Science Publications, 1995,
- ↑ W. Kutzelnigg, Phys. Scr. 1987, 36, 416–431.
- ↑ Blixt, J.; Glaser, J.; Mink, J.; Persson, I.; Persson, P.; Sandstroem, M.; J. Am. Chem. Soc.; 1995, 117, 5089-5104. DOI:10.1021/ja00123a011
- ↑ http://www.huntresearchgroup.org.uk/teaching/teaching_comp_lab_year3/4c_mixed_pp_basis.html
- ↑ The Molecular Structures of Boron Trimethyl, Trifluoride, Trichloride, and Tribromide. Henri A. Lévy, L. O. Brockway, J. Am. Chem. Soc., 1937, 59 (11), pp 2090.
- ↑ Generalization of Natural Bond Orbital Analysis to Periodic Systems: Applications to Solids and Surfaces via Plane-Wave Density Functional Theory. Benjamin D. Dunnington and J. R. Schmidt, J. Chem. Theory Comput., 2012, 8 (6), pp 1902–1911. DOI:10.1021/ct300002t
- ↑ Glendening, E. D.; Landis, C. R.; Weinhold, C.; Advanced Review, 2011, 2, 1-42. DOI:10.1002/wcms.51
- ↑ Iron Complex-Catalyzed Ammonia–Borane Dehydrogenation. A Potential Route toward B–N-Containing Polymer Motifs Using Earth-Abundant Metal Catalysts. R. Tom Baker, John C. Gordon, Charles W. Hamilton, Neil J. Henson, Po-Heng Lin, Steven Maguire, Muralee Murugesu, Brian L. Scott, and Nathan C. Smythe. J. Am. Chem. Soc., 2012, 134 (12), pp 5598–5609. DOI: 10.1021/ja210542r
- ↑ S. Xie, Y. Song, and Z. Liu, Can. J. Chem. 87, 1235–1247 (2009).