Rep:Mod2:LZT11218
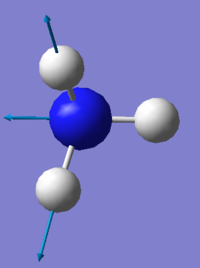
NH3 molecule
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP): -56.55776873a.u.
RMS gradient: 0.00000485a.u.
Point group: C3V
N-H bond length= 1.01798Å ( ±0.01Å )
N-H bond angle= 105.741° ( ±1° )
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986260D-10
The optimisation file is linked to here
NH3 |
NH3 molecule's vibration modes
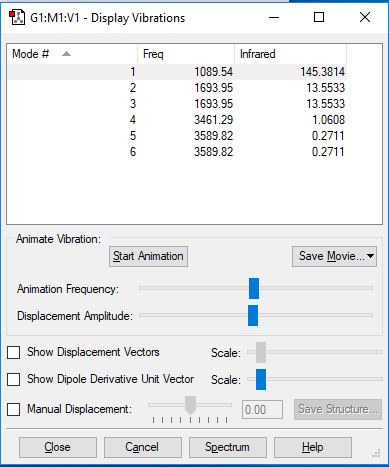
| Motion(vectors) | mode1 | mode2 | mode3 | mode4 | mode5 | mode6 |
|---|---|---|---|---|---|---|
| Image | 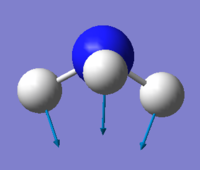 |
 |
 |
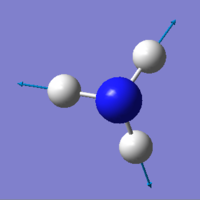 |
 |
|
| wavenumber/cm−1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity (arbitrary units) | 145 | 14 | 14 | 1 | 0 | 0 |
Number of mode expected: 6
In this case the second and the third mode, the fifth and the sixth mode are degenerated.
Mode 1,2,3 are bending; Mode 4,5,6 are stretching.
Mode 4 is higly symmetrical.
Mode 1 is known as "umbrella" mode.
Three bands are expected to see, which is mode 1, 2 and 3.
NH3 molecule's charge distribution
The charged on N atom is -1.125, on H atom is +0.375. In experiment, we expected their charge varies because the electron will move around and the charge distribution will change all the time.
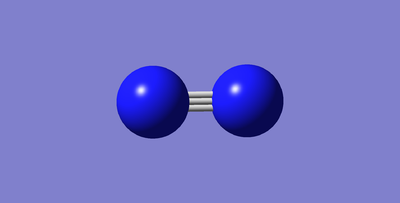
N2 and H2 molecular
N2
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP): -109.52359111a.u.
RMS gradient: 0.02473091a.u.
Point group: Dinfinity
Nitrogen triple bond bond length= 1.10550Å ( ±0.01Å )
Nitrogen triple bond Bond angle= 180° ( ±1° )
The optimisation file is linked to here
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401081D-13
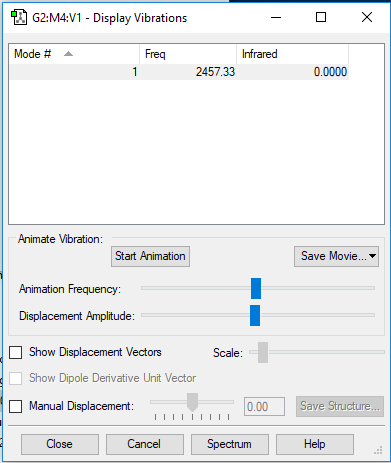
Vibration:
| wavenumber/cm−1 | 2457 |
|---|---|
| symmetry | SGG |
| intensity (arbitrary units) | 0 |
No dipole change in this case so no band show at IR spectrum.
The charge on both atom are zero as they are identical.
N2 |
H2
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP): -1.17853936a.u.
RMS gradient: 0.00000017a.u.
Point group: Dinfinity
H-H bond length= 0.74279Å ( ±0.01Å )
H-H bond angle= 180° ( ±1° )

The optimisation file is linked to here
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13
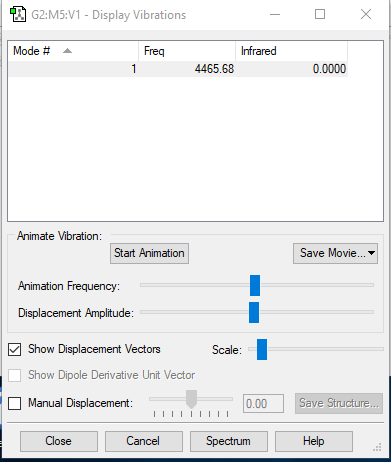
Vibration:
| wavenumber/cm−1 | 4466 |
| symmetry | SGG |
| intensity (arbitrary units) | 0 |
No dipole change in this case so no band show at IR spectrum.
The charge on both atom are zero as they are identical.
H2 |
Experimental data of N2
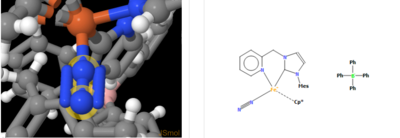
Thea link of mono-metallic TM complex that coordinates N2 [1]
Identifier name: VEJDIA
Nitrogen bond length: 1.105Å
This is the same as the computational value.In the metal complex, the NN triple bond are bonded to Fe-, the triple bond keep the same it contains the same property the bond here might be formed by two valance electrons from the Fe- the might be the reason that the middle N atom form 4 bonds and keep the same property(the triple bond's length does not change).
Reaction and energy
N2 + 3H2-----2NH3
E(NH3)= -56.55776873a.u.
2*E(NH3)= -113.11553746a.u.
E(N2)= -109.52359111a.u.
E(H2)= -1.17853936a.u.
3*E(H2)= -3.53561808a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05632827a.u. = -147.889873kJ/mol
The ammonia product is more stable.
NF3
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP): -354.07131058a.u.
RMS gradient: 0.00010256a.u.
Point group: C3V
N-F bond length= 1.38404Å ( ±0.01Å )
N-F bond angle= 101.830° ( ±1° )

Item Value Threshold Converged? Maximum Force 0.000164 0.000450 YES RMS Force 0.000108 0.000300 YES Maximum Displacement 0.000612 0.001800 YES RMS Displacement 0.000296 0.001200 YES Predicted change in Energy=-1.274066D-07
The optimisation file is linked to here
NF3 |
| Motion(vectors) | mode1 | mode2 | mode3 | mode4 | mode5 | mode6 |
| Image |  |
 |
 |
 |
 |

|
| wavenumber/cm−1 | 482 | 482 | 644 | 930 | 930 | 1062 |
| symmetry | E | E | A1 | E | E | A1 |
| intensity (arbitrary units) | 1 | 1 | 3 | 208 | 208 | 40 |
Number of mode expected: 6
Mode 3 and 6 are the most symmetrical.
Mode 1 and 2; 4 and 5 are degenerated.
Three bands are expected to see, which is mode 4, 5 and 6.
The charged on N atom is 0.636, on F atom is -0.212
Reference
Knowledge of MO orbitals are all from lecture Chemical Bonding. All diagram are took by screenshot from Gaussian.
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES - however, correctly stated that there are two sets of degenerate modes - this explains a spectrum with 4 peaks. However there are only 2 peaks visible as peaks 4, 5 and 6 are of too low an intensity to be visible. Therefore, the number of 3 bands you quoted is incorrect.
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES - however, you stated a bond angle for diatomic molecules. To define a bond angle a minimum of 3 atoms is needed!
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
NO - Your value is close to the real one of 146.5kJ/mol. Probably this difference is only a result of different conversion factors or rounding the intermediate results incorrectly.
Have you reported your answers to the correct number of decimal places?
NO - the energy in kJ/mol should only be reported to one decimal place!
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You could have explained the charges using an electronegativity argument. You stated to expect 3 bands in the experimental spectrum. Two of the modes you stated to be responsible for the bands are degenerate. Therefore, only 2 bands should be seen. You missed to explain if the MOs you are displaying are occupied or unoccupied. You missed to mention if the HOMO and LUMO are bonding/anti-bonding or non-bonding.Additionally you missed to mention the contributing AOs for the HOMO and LUMO. You correctly stated the contributing AOs for the other MOs. You are confused about the meaning of bonding/anti-bonding and non-bonding! The very same MO cannot be bonding and anti-bonding at the same time. (MO5 and 6). A node does not necessarily mean it is a anti-bonding MO (MO3, this is actually a non-bonding one)
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far
NO - No independent work has been identified.