Rep:MOD:ZJY0309
NH3
Optimisation
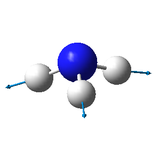
Molecule: NH3
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP) (au): -56.55777
RMS gradient (au): 0.00000323
Point group: C3V
N-H bond distance: 1.02Å
H-N-H bond angle: 105.7°
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000014 0.001800 YES RMS Displacement 0.000009 0.001200 YES
Jmol dynamic image of NH3 |
The optimisation file is liked to here
Frequency Analysis
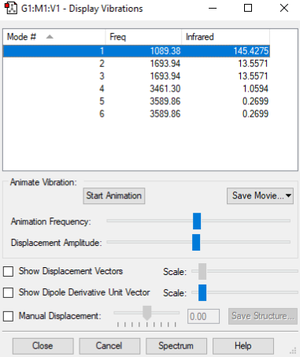
| wavenumber cm-1 | 1089 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity arbitrary units | 145 | 14 | 14 | 1 | 0 | 0 |
| image |  |
 |
 |
 |
 |
|
How many modes do you expect from the 3N-6 rule?
6
Which modes are degenerate (ie have the same energy)?
2 modes with frequency of 1694 cm-1 and 2 modes with frequency of 3590 cm-1
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
bending vibration: 1089 cm-1, 1694 cm-1, 1694 cm-1
bond stretch vibration: 3461 cm-1, 3590 cm-1, 3590 cm-1
Which mode is highly symmetric?
3461 cm-1 and 1089 cm-1
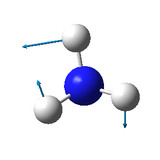
One mode is known as the "umbrella" mode, which one is this?
1089 cm-1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2. Theoretically there should be 4 bands due to 2 degenerate modes, but due to the low intensity of the bond stretch vibration (rel. intensity=1 and 0), 2 bond stretch bands cannot be seen in the experimental spectrum.
Charge Analysis
| N-atom | H-atoms | image |
|---|---|---|
| -1.125 | 0.375 | 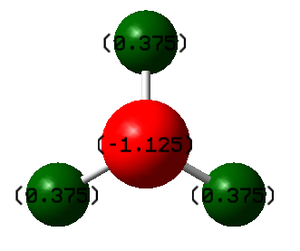
|
It is expected that N-atom should have a partially negative charge whereas H-atoms should have partially positive charges, because nitrogen is more electronegative than hydrogen so the electron density on N-atom is higher than the one on H-atoms.
N2
Optimisation
Molecule: N2
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP) (au): -109.52413
RMS gradient (au): 0.00000060
Point group: N∞h
N-N bond distance: 1.11Å
For lack of 3 atoms, the bond angle cannot be calculated.
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Jmol dynamic image of N2 |
The optimisation file is liked to here
Frequency Analysis
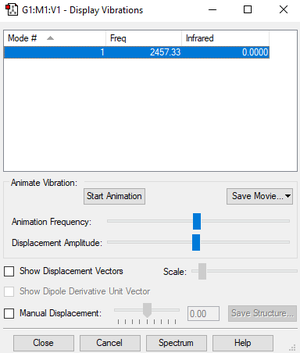
| wavenumber cm-1 | 2457 |
| symmetry | SGG |
| intensity arbitrary units | 0 |
| image | 
|
Charge Analysis
| N-atom | N-atom |
|---|---|
| 0.000 | 0.000 |
It is expected that the atoms should have no partial charge because 2 same atoms have the same elctronegativities.
H2
Optimisation
Molecule: H2
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP) (au): -1.17854
RMS gradient (au): 0.00000017
Point group: N∞h
H-H bond distance: 0.74Å
For lack of 3 atoms, the bond angle cannot be calculated.
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Jmol dynamic image of H2 |
The optimisation file is liked to here
Frequency Analysis

| wavenumber cm-1 | 4466 |
| symmetry | SGG |
| intensity arbitrary units | 0 |
| image | 
|
Charge Analysis
| H-atom | H-atom |
|---|---|
| 0.000 | 0.000 |
It is expected that the atoms should have no partial charge because 2 same atoms have the same elctronegativities.
Crystal Structure Comparison
The CDDC link to the molecule can be found at:[[1]]
The bond length (1.09Å) is slightly shorter the optimised one (1.11Å) due to the following reasons:
1. From the computational perspective, Gaussian calculates the bond length of the molecule in gas phase, while the experimental bond length is measured in the crystalline structure of the molecule; therefore, two bond lengths will be different.
2. From the experimental perspective, it can be seen from the 3D structure of this molecule that N≡N is a bit stretched and exposed to the outside of the whole molecule; so, due to packing effects of the crystal structure, the bond length may be shortened.
Haber-Bosch Process Energy Calculation
ENH3= -148492.43 kJ/mol
2*ENH3= -296984.86 kJ/mol
EN2= -287555.62 kJ/mol
EH2= -3094.26 kJ/mol
3*EH2= -9282.78 kJ/mol
ΔE=2*ENH3-[EN2+3*EH2]= -146.46 kJ/mol
Ammonia product is more stable than the reactants because the reaction is exothermic and releases energy (i.e. The energy of the reactants is greater than that of the product.).
[CN]-
Optimisation
Molecule: [CN]-
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP) (au): -92.82453
RMS gradient (au): 0.00000704
Point group: C∞v
C-N bond distance: 1.18Å
For lack of 3 atoms, the bond angle cannot be calculated.
Item Value Threshold Converged? Maximum Force 0.000012 0.000450 YES RMS Force 0.000012 0.000300 YES Maximum Displacement 0.000005 0.001800 YES RMS Displacement 0.000008 0.001200 YES
Jmol dynamic image of [CN]- |
The optimisation file is liked to here
Frequency Analysis
| wavenumber cm-1 | 2139 |
| symmetry | SG |
| intensity arbitrary units | 8 |
| image | 
|
Charge Analysis
| N-atom | C-atom | image |
|---|---|---|
| -0.754 | -0.246 | 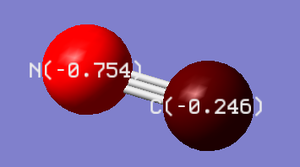
|
It is expected that N-atom should possess more negative charge than C-atom, for nitrogen is more electronegative than carbon. This charge distribution also converges with the overall charge being -1.
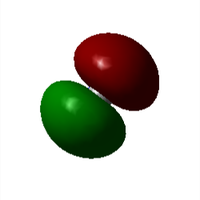
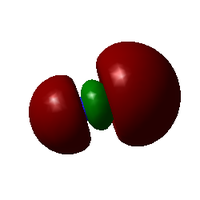
Molecular Orbitals
Further Analysis
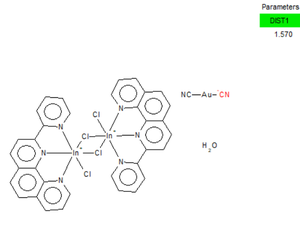
The CDDC link to the molecule can be found at:[[2]]
The bond length (1.57Å) is much longer than the optimised one (1.18Å) probably due to the following reasons:
1. From the computational perspective, Gaussian calculates the bond length of the molecule in gas phase, while the experimental bond length is measured in the crystalline structure of the molecule; therefore, two bond lengths will be different.
2. From the experimental perspective, due to the strong interactions between gold and [CN]-, the π* orbital in [CN]-, for instance, may overlap with the dxy orbital in Au; however, backbonding may also exist when the dz2 overlaps with pz orbital in [CN]- due to symmetry, which may reduce the bond strength between Au and [CN]-.
It is interesting that although [CN]- and N2 are isoelctronic, but their bonding behaviour can be very different in different complexes.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
NO - Energies in kJ/mol should only be reported up to one decimal place.
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES
You could have discussed the calculated vibrational mode The second displayed MO is a bonding one rather than an anti-bonding. The last displayed MO is not only in the HOMO region, ti is the HOMO of CN-. You could have explained the energetic order of the MOs in more detail (number of nodes...).
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
YES
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far