Rep:MOD:YZ20215
NH3 Molecule
The optimisation file is linked to here
General Information
Optimisation is done and seven intermediates were shown with the final result below:
N-H bond distance/Angstroms=1.01798
H-N-H bond angle/degrees=105.741
File Name: yanzhang_nh3_optf
Molecule Name: NH3
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP): -56.55776873 a.u.
Point Group: C3V
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
test molecule |
Vibrations
Questions 1.how many modes do you expect from the 3N-6 rule?
3*4-6=6
2.which modes are degenerate (ie have the same energy)?
5 and 6
3.which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending:1,2,3 Bond stretch:4,5,6
4.which mode is highly symmetric?
4
5.one mode is known as the "umbrella" mode, which one is this?
1
6.how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2, because there are N-H bending and streching.
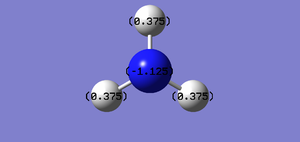
Charge
Charge on N atom=-1.125
Charge on H atom=0.375
N atom is expected to have negative as it is much more electronegative than hydrogen atoms, and the H atoms are expected to have positive charge.
H2 Molecule
The optimisation file is linked to here
test molecule |
General Information
File Name: yz20215_H2_OPTF
Molecule Name: H2
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
as there is no overall polarity, the charge on both atoms is zero
Spin: Singlet
E(RB3LYP): -1.17853936 a.u.
RMS Gradient Norm: 0.00000017 a.u.
Imaginary Freq: 0
Dipole Moment: 0.0000 Debye
Point Group: D∞H
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Vibrations
1 vibration mode is expected as 3N-5 rules apply to linear molecules.
N2 Molecule
The optimisation file is linked to here
test molecule |
General Information
File Name: yz20215_N2_OPTF
Molecule Name: N2
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
as there is no overall polarity, the charge on both atoms is zero
Spin:Singlet
E(RB3LYP): -109.52412868 a.u.
RMS Gradient Norm: 0.00000060 a.u.
Imaginary Freq: 0
Dipole Moment: 0.0000 Debye
Point Group:D∞H
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Vibrations
1 vibration mode is expected as 3N-5 rules apply to linear molecules.
Haber-Bosch Reaction Energy
E(NH3)=-56.55776873 a.u.
2*E(NH3)=-113.11553746 a.u.
E(N2)=-109.52412868 a.u.
E(H2)=-1.17853936 a.u.
3*E(H2)=-3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.0557907 a.u.=-146.47849401 kJ/mol
The process is exothermic, indicating that the ammonia molecule is more stable.
Literature value=-92.4 kJ/mol
The value calculated deriviates largely from the literature value, so the method using the software to obtained the energy is not very accurate.
Project Molecule: S2 Molecule
The optimisation file is linked to here
test molecule |
General Information
File Name: YZ20215_S2_OPTF
Molecule Name: S2
S=S double bond distance/Angstroms=1.77760
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0 on both atoms
as there is no overall polarity, the charge on both atoms is zero
Spin: Singlet
E(RB3LYP): -796.32599779 a.u.
RMS Gradient Norm: 0.00000372 a.u.
Imaginary Freq: 0
Dipole Moment: 0.0000 Debye
Point Group: D∞H
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000011 0.001800 YES RMS Displacement 0.000016 0.001200 YES
Vibrations
1 vibration mode is expected as 3N-5 rules apply to linear molecules.
Molecular Orbital Diagrams
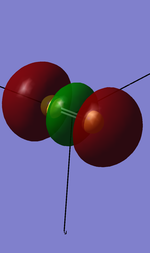
1. 3s sigma* u antibonding orbital
Components: two 3s valence shell atomic orbitals from both S atoms
Character: ocuppied antibonding orbital with no mixing
Energy: -0.61535 a.u.
High in energy compared with 2s,2p,and 1s orbitals, but the second lowest in the valence shell orbitals.
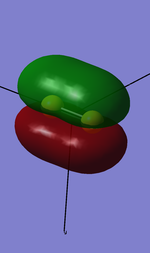
2. 3p sigma g bonding orbital(in z direction)
Components: two 3pz valence shell atomic orbitals from both S atoms overlap head-to-head
Character: ocuppied bonding orbital with mixing with 3s sigma orbital
Energy: -0.39516 a.u.
High in energy compared with 2s,2p,and 1s orbitals, but the third lowest in the valence shell orbitals.
The red lobes are larger due to mixing.
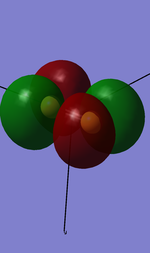
3. 3p pi u bonding orbital(in y direction)
Components: two 3py valence shell atomic orbitals from both S atoms
Character: ocuppied bonding orbital with no mixing
Energy: -0.36284 a.u.
High in energy compared with 2s,2p,and 1s orbitals, but the third highest in the valence shell orbitals.
4. 3p pi u bonding orbital(in x direction)
Components: two 3px valence shell atomic orbitals from both S atoms
Character: ocuppied bonding orbital with no mixing
Energy: -0.34959 a.u.
High in energy compared with 2s,2p,and 1s orbitals, the second highest in the valence shell orbitals.
5. 3p pi* u antibonding orbital(in x direction)
Components: two 3px valence shell atomic orbitals from both S atoms
Character: ocuppied bonding orbital with no mixing, HOMO
Energy: -0.21842 a.u.
Highest in energy compared with 2s,2p,and 1s orbitals.