Rep:MOD:PT1118
NH3 molecule
The first molecule I looked at was NH3 .
Calculation Summary
Molecule = NH3
Calculation Method = RB3LYP
Basis set = 6-31G(d,p)
Final energy E(RB3LYP) = -56.55776873 a.u.
RMS Gradient Norm = 0.00000485 a.u.
Point Group = C3V
N-H bond length = 1.02 Å (accurate to ≈ 0.01Å)
H-N-H bond angle = 106° (accurate to ≈ 1°)
Item table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Jmol Image
Ammonia molecule |
The optimisation file is linked to here
Vibrations Analysis
| symmetry | wavenumber (cm-1) | intensity of vibration (arbitrary units) | image |
|---|---|---|---|
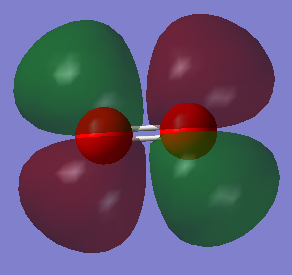
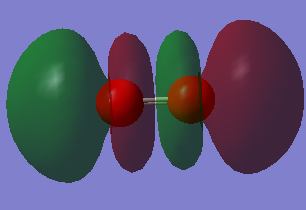
| A1 | 1090 | 145 | 
|
| E | 1694 | 14 | 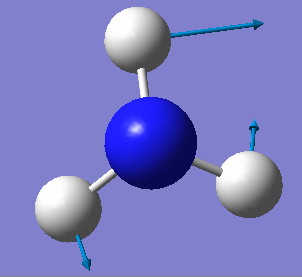
|
| E | 1694 | 14 | 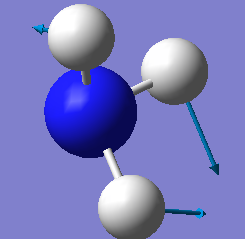
|
| A1 | 3461 | 1 | 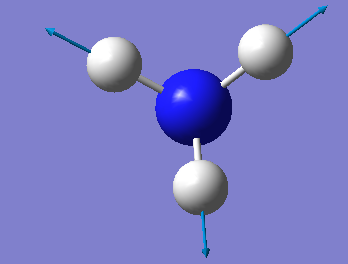
|
| E | 3590 | 0 | 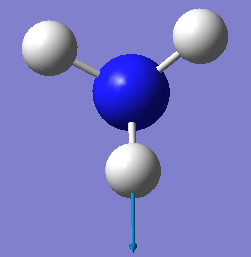
|
| E | 3590 | 0 | 
|
6 modes are expected.
There is 2 sets of modes that are degenerate, the vibrations 1694 cm-1 and 3590cm-1.
Vibrations 1090cm-1, 1694cm-1 and 1694cm-1 are bending vibrations and vibrations 3461cm-1, 3590cm-1 and 3590cm-1 are bond stretch vibrations.
Vibration 3461cm-1 is highly symmetric.
Vibration 1090cm-1 is known as the "umbrella" mode.
You expect 3 bands from 1090cm-1, 1694cm-1 and 1694cm-1 in an experiment spectrum of gaseous ammonia, but 2 are degenerate and so you expect to just see 1 peak for them. Therefore 2 bands are seen.
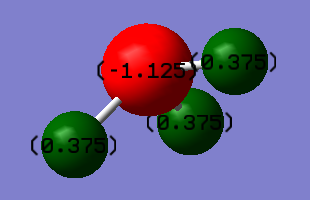
Charge Analysis
The charge of the N atom is -1.125 and the charge of the hydrogen atoms are 0.375. Nitrogen is more electronegative than hydrogen. Therefore, nitrogen pulls more electron density towards it and so I expect a negative charge on nitrogen and positive charge on hydrogen.
N2molecule
The next molecule analysed was N2.
Calculation Summary
Molecule = N2
Calculation Method = RB3LYP
Basis set = 6-31G(d,p)
Final energy E(RB3LYP) = -109.52412868 a.u.
RMS Gradient Norm = 0.00000060 a.u.
Point Group = D*H
N-N bond length = 1.11 Å (accurate to ≈ 0.01Å)
Item table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Jmol Image
nitrogen molecule |
The optimisation file is linked to here
Vibrations Analysis
| symmetry | wavenumber (cm-1) | intensity of vibration (arbitrary units) | image |
|---|---|---|---|
| SGG | 2457 | 0 | 
|
1 mode is expected. 2457cm-1 is a bond stretch vibration. You would expect to see no band in an experimental spectrum of gaseous nitrogen because there is no change in dipole moment.
Charge Analysis
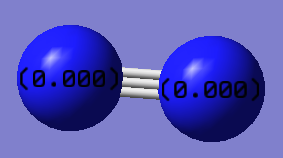
The charge of both the N atoms is 0, this is because it is a symmetric molecule.
H2molecule
The next molecule analysed was H2.
Calculation Summary
Molecule = H2
Calculation Method = RB3LYP
Basis set = 6-31G(d,p)
Final energy E(RB3LYP) = -1.17853936 a.u.
RMS Gradient Norm = 0.00000017 a.u.
Point Group = D*H
H-H bond length = 0.74 Å (accurate to ≈ 0.01Å)
Item table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Jmol Image
Hydrogen molecule |
The optimisation file is linked to here
Vibrations Analysis
| symmetry | wavenumber (cm-1) | intensity of vibration (arbitrary units) | image |
|---|---|---|---|
| SGG | 4466 | 0 | 
|
1 mode is expected. 4466 cm-1 is a bond stretching vibration. No band is expected to be seen in an experimental spectrum of gaseous hydrogen because there is no change in dipole moment.
Charge Analysis
The charge of both the H atoms is 0 because it is a symmetric molecule.
Transition metal complex
The structure I looked at was DAMSUG. The two N-N bond distances are 1.128Å and 1.134Å in the structure. The bond length I obtained is 1.10550Å. Therefore, the N-N bond in the molecule is longer than the N-N bond measured computationally, and so the bond is weaker. The bond is longer because when it bonds to the transition metal, the transition metal attracts electron density from the nitrogen and so weakening and lengthening the N-N bond. Due to it being measured computationally, then if a different computational method was used the bond length may change.
The structure is linked [1]
The Haber-Bosch process
N2 + 3H2 -> 2NH3
Energies
E(NH3)= -56.5577687 a.u.
2*E(NH3)= -113.115538 a.u.
E(N2)= -109.5241287 a.u.=-287555.6 kJ mol-1
E(H2)= -1.1785394 a.u.= -3094.3 kJ mol-1
3*E(H2)= -3.5356181 a.u.
ΔE= 2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557907 a.u.= -146.5 kJ mol-1
The product is more stable.
O2 molecule
The molecule of my choice is O2 molecule.
Calculation Summary
Molecule = O2
Calculation Method = RB3LYP
Basis set = 6-31G(d,p)
Final energy E(RB3LYP) = -150.25742434 a.u.
RMS Gradient Norm = 0.00007502 a.u.
Point Group = D*H
O-O bond length = 1.22 Å (accurate to ≈ 0.01Å)
Item table
Item Value Threshold Converged? Maximum Force 0.000130 0.000450 YES RMS Force 0.000130 0.000300 YES Maximum Displacement 0.000080 0.001800 YES RMS Displacement 0.000113 0.001200 YES
Jmol Image
Ammonia molecule |
The optimisation file is linked to here
Vibrations Analysis
| symmetry | wavenumber (cm-1) | intensity of vibration (arbitrary units) | image |
|---|---|---|---|
| SGG | 1643 | 0 | 
|
1 mode is expected. 1643 cm-1 is a bond stretch vibration. You would expect to see no band in an experimental spectrum of gaseous oxygen because there is no change in dipole moment.
Charge Analysis
The charge of both the O atoms is 0, this is because the bond is between two atoms of the same element and so the electron density is shared equally. It is a symmetrical molecule.
O2 molecular orbitals
I chose to look at molecular orbitals (MO) 1, 4, 8, 9 and 10.
MO 1 has an energy of -19.30736 a.u.. It forms the 1σg MO. It is a bonding MO that is formed from a 1s atomic orbital (AO) from each oxygen atom and so this MO is occupied. The MO is deep in energy because the 1s electrons are close to the nucleus due to the strong electrostatic attractions between the nucleus and electron and therefore do not overlap.
MO 4 an energy of -0.79821 a.u. It forms an antibonding MO, 2σu*. A 2s AO from each oxygen atom contributes to form this. The s orbitals have different phases and have a node between them and so do not overlap. This MO is occupied. It is deep in energy and so not involved in bonding.
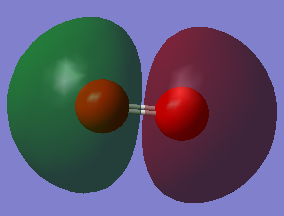
MO 8 is the HOMO, therefore this MO is occupied. It has an energy of -0.25018 a.u.. This is an antibonding MO, 1πg*. It can be formed from 2px or 2pz AO. This orbital is still low in energy but it is not as low as the MO 1 and so the electrons are more available for bonding. It forms an antibonding MO and so this destabilises the pi bond.
MO 9 is the LUMO and so this is unoccupied. It has an energy of -0.17928 a.u.. It could be formed from 2px or 2pz AOs. This is an antibonding MO, 1πg*. When bonding this level will accept electrons. It is higher in energy than the HOMO.
MO 10 has an energy of 0.21200 a.u.. 2px AO contribute to this MO. This is an antibonding MO, 3σu*. It 3 nodes and so that is why it has high energy.
H2O Molecule
(independence mark)
Calculation Summary
Molecule = H2O
Calculation Method = RB3LYP
Basis set = 6-31G(d,p)
Final energy E(RB3LYP) = -74.41973740 a.u.
RMS Gradient Norm = 0.00006276 a.u.
Point Group = C2V
O-H bond length = 0.97 Å (accurate to ≈ 0.01Å)
H-O-H bond angle = 104° (accurate to ≈ 1°)
Item table
Item Value Threshold Converged? Maximum Force 0.000099 0.000450 YES RMS Force 0.000081 0.000300 YES Maximum Displacement 0.000115 0.001800 YES RMS Displacement 0.000120 0.001200 YES
Jmol Image
Ammonia molecule |
The optimisation file is linked to here
Vibrations Analysis
| symmetry | wavenumber (cm-1) | intensity of vibration (arbitrary units) | image |
|---|---|---|---|
| A1 | 1665 | 70 | 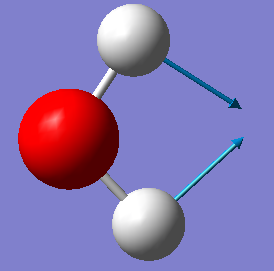
|
| A1 | 3801 | 2 | 
|
| B2 | 3914 | 20 | 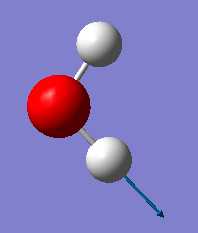
|
3 modes are expected. 3801 cm-1 and 3914cm-1 are bond stretching vibrations. 2 bands are expected to be seen in an experimental spectrum of gaseous H2O.
Charge Analysis
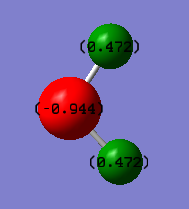
The charge of both the hydrogen atoms are 0.472 and the charge of the oxygen atom is -0.944. This is because oxygen is more electronegative than hydrogen and so it attracts more electron density causing the charge to be negative.
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES - however you used the same label ('ammonia molecule' for a bunch of jmols which are definitely not showing NH3.
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You partially missed to state if the MOs are occupied/unoccupied (MOs 10) and you stated MO 1 to be bonding. You correctly stated that no overlap is observed between the 1s AOs of the Os. Therefore this MO is called non-bonding even though the AOs have the phase they need to have for a bonding MO.
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or
YES - well done!
Do some deeper analysis on your results so far