Rep:MOD:01372542
NH3 Molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -56.55776873 au
RMS Gradient: 0.00000485 au
Point group: C3V
N-H Bond Length: 1.01798 Å
H-N-H Bond Angle: 105.741o
Item Table For NH3:
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986282D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
NH3 3D Structure |
The optimisation file for NH3 is linked to here
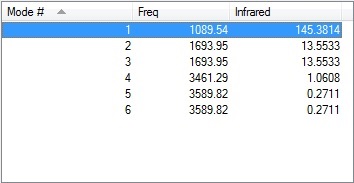
1) How many modes do you expect from the 3N-6 rule?
As there are 4 atoms in an NH3 molecule, by the 3N-6 rule you would expect to get 6 nodes (as 3(4)-6=6).
2) Which modes are degenerate?
Modes 2 and 3, and 5 and 6 form degenerate pairs, shown by their identical vibrational frequencies.
3) Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Modes 1,2 and 3 are bending vibrations, and modes 4,5 and 6 are formed from bond stretch vibrations.
4) Which mode is highly symmetric?
Mode 4 is highly symmetric
5) One mode is known as the "umbrella" mode, which one is this?
Mode 1 resembles the opening and closing mechanism of an umbrella.
6) How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
You would expect to see 2 bands in a spectrum of gaseous ammonia, the first from mode 1 and the second from modes 2 and 3 combined as they occur at the same frequency. Modes 4,5 and 6 have intensities that are far too low to be visible on the spectrum, due to the symmetric stretches and small changes in the dipole moment. In reality the spectrum has a third peak[1], in the region of 3500, potentially caused by the combination of smaller vibrations.
Atomic Charges
Within the molecule, the nitrogen has a charge of -1.125, and each hydrogen has a charge of 0.375. This agrees with what I would expect, as nitrogen is the more electronegative atom and will pull the bonded pair of electrons towards itself, increasing its electron density and therefore reducing its charge.
N2 Molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -109.52412868 au
RMS Gradient: 0.00000060 au
Point group: D∞h
N-N Bond Length: 1.10550 Å
Item Table For N2:
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400969D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
N2 3D Structure |
The optimisation file for N2 is linked to here
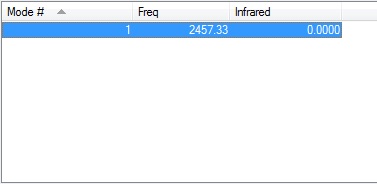
H2 Molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -1.17853936 au
RMS Gradient: 0.00000017 au
Point group: D∞h
H-H Bond Length: 0.74279 Å
Item Table For H2:
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
H2 3D Structure |
The optimisation file for H2 is linked to here
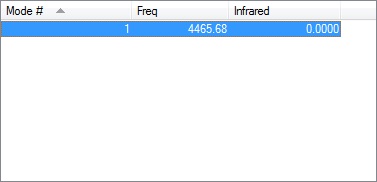
Energy of Reaction
E(NH3) = -56.55776873 au
2*E(NH3) = -113.1155375 au
E(N2) = -109.5241287 au
E(H2) = -1.17853936 au
3*E(H2) = -3.53561808 au
ΔE = 2*E(NH3)-(E(N2)+3*E(H2)) = -0.0557907 au = -146.48 kJ/mol From this we can tell that the product, ammonia, is more stable than the reactants N2 and H2.
In literature, this value was cited as -99.22 kJ/mol [2]. The difference between this value and the computed value is due to poor optimisation of the molecule.
CF4 Molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -437.47627267 au
RMS Gradient: 0.00004049 au
Point group: TD
C-F Bond Length: 1.32939 Å
Item Table For CF4:
Item Value Threshold Converged?
Maximum Force 0.000078 0.000450 YES
RMS Force 0.000042 0.000300 YES
Maximum Displacement 0.000133 0.001800 YES
RMS Displacement 0.000071 0.001200 YES
Predicted change in Energy=-2.081896D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3294 -DE/DX = -0.0001 !
! R2 R(1,3) 1.3294 -DE/DX = -0.0001 !
! R3 R(1,4) 1.3294 -DE/DX = -0.0001 !
! R4 R(1,5) 1.3294 -DE/DX = -0.0001 !
! A1 A(2,1,3) 109.4712 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4712 -DE/DX = 0.0 !
! A3 A(2,1,5) 109.4712 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.4712 -DE/DX = 0.0 !
! A5 A(3,1,5) 109.4712 -DE/DX = 0.0 !
! A6 A(4,1,5) 109.4712 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) -120.0 -DE/DX = 0.0 !
! D3 D(2,1,5,4) 120.0 -DE/DX = 0.0 !
! D4 D(3,1,5,4) -120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
CF4 3D Structure |
The optimisation file for CF4 is linked to here
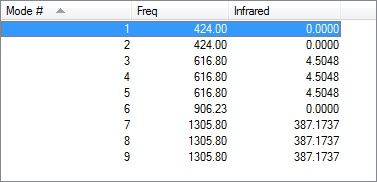

CF4 Molecular Orbitals

This first molecular orbital is a bonding orbital, formed from the overlap of 2s sub-shells from both the central carbon atom and the surrounding fluorine atoms. There is a very large overlap of these orbitals as they hold valence electrons and are therefore heavily involved in bonding. Evidently this orbital is also occupied.

This molecular orbital is formed from all 2p orbitals, shown by the nodes on each atom. There is favourable overlap over all bonds showing that this is a bonding MO. This orbital is occupied, and does not have a particularly deep energy, so it will contribute fairly significantly to the overall bond. The three p orbitals on carbon could be involved in this MO, meaning it has three possible orientations, forming three degenerate MOs.

MO number three is formed from 2s orbitals from both the carbon and fluorine, however, they are out of phase this time and therefore form the antibonding orbital, shown by the node on each bond. The orbital is fully occupied, and not particularly deep in energy, meaning it will have some influence on the bonding of the molecule. However, it does not lie near the LUMO/HOMO region and does not carry valence electrons so its impact on the strength of the bond is limited.

This MO is comprised of a 2p orbital on the carbon, and s orbitals on the surrounding fluorine. Of the fluorine atoms, two are in one phase and two are in the other, with each pair lining up with the corresponding end of the carbon's p orbital. The overlap is in phase across the whole bond for each C-F bond, showing that the molecule is a bonding MO. The p orbital can be in 3 possible orientations so there are three degenerate MOs that look the same, each with a fairly low energy. As a result of the low energy it is unlikely that this contributes much to the overall bonding of the molecule.

This filled MO is very low in energy, and it is formed of a 2s sub shell from the carbon. This is a non-bonding orbital, shown by the lack of influence from any adjacent fluorine atoms. It has no influence on bonding as it is not a bonding or antibonding orbital.
CF4 Charges

The central carbon atom is very electropositive due to the surrounding fluorine atoms drawing away the bonded electrons. Fluorine atoms are very electronegative so will attract the bonded pair of electrons to itself, shifting the negative charge from the carbon to towards the fluorine.
References
- ↑ http://webbook.nist.gov/cgi/cbook.cgi?ID=C7664417&Type=IR-SPEC&Index=1.
- ↑ Ebbing, Darrel D.General Chemistry 3rd ed.;Houghton Mifflin Company: Boston, MA, 1990 pp 115,175, 223-4, 227.
