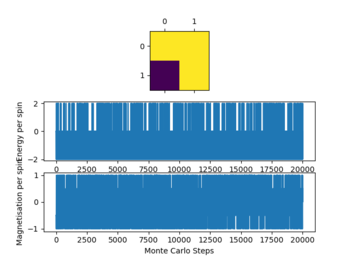
Rep:LH3115-Montecarlo
Montecarlo experiment
Introduction to the Ising model
TASK: 1
The lowest energy is achieved if spins align, therefore and so as spins are either -1 or +1.
Therefore our equation becomes:
As the total number of neighbors is the expression And the sum of all atoms if every is is just N times the number of interactions, which is .
So the overall expression is:
The multiplicity of the state is 2 as there are two possible states, all parallel with spins all up or all down.
so here
TASK: 2
as in 3 dimensions the atom has 6 neighbours that will change their interaction with it.
The entropy gain depends on the number of particles, if one particle flips the entropy gain is Where goes from 1 to 1000 because , therefore
TASK: 3
For both examples provided the magnetisation M = 1:
For the 1D lattice of 5:
For the 2d lattice of 25:
At absolute 0 the system will adapt the lowest energy configuration thus all spins will be parallel. This will give a magnetisation
Calculating the energy and magnetisation
Task 4 and Task 7
Code for the initial IsingLattice function and the Montecarlostep
import numpy as np
class IsingLattice:
E = 0.0
E2 = 0.0
M = 0.0
M2 = 0.0
n_cycles = 0
def __init__(self, n_rows, n_cols):
self.n_rows = n_rows
self.n_cols = n_cols
self.lattice = np.random.choice([-1,1], size=(n_rows, n_cols))
def energy(self):
"Return the total energy of the current lattice configuration."
sumpart=0
for i in range(self.n_rows):
i1 = (i+1)%(self.n_rows)
i2 = (i-1)%(self.n_rows)
for ic in range(self.n_cols):
ic1 = (ic+1)%(self.n_cols)
ic2 = (ic-1)%(self.n_cols)
sumpart = sumpart + self.lattice[i][ic] * self.lattice[i1][ic]
sumpart = sumpart + self.lattice[i][ic] * self.lattice[i2][ic]
sumpart = sumpart + self.lattice[i][ic] * self.lattice[i][ic1]
sumpart = sumpart + self.lattice[i][ic] * self.lattice[i][ic2]
energy = (-0.5)*sumpart
return energy
def magnetisation(self):
"Return the total magnetisation of the current lattice configuration."
sumspin = 0
for i in self.lattice:
for j in i:
sumspin = sumspin+j
magnetisation=sumspin
return magnetisation
def montecarlostep(self, T):
# complete this function so that it performs a single Monte Carlo step
energy = self.energy()
#the following two lines will select the coordinates of the random spin for you
random_i = np.random.choice(range(0, self.n_rows))
random_j = np.random.choice(range(0, self.n_cols))
#the following line will choose a random number in the rang e[0,1) for you
random_number = np.random.random()
#change spin of random number
self.lattice[random_i,random_j] = -self.lattice[random_i,random_j]
#find delta E
newe=self.energy()
deltae=newe-energy
#determine whether new values can be accepted straigth away, no else function needed as the values are already changed
if deltae > 0:
expocalc=np.exp(-deltae/T)
if random_number>expocalc:
#change lattice back as it below R
self.lattice[random_i,random_j] = -self.lattice[random_i,random_j]
energy=self.energy()
magnetisation=self.magnetisation()
#summations of values
self.n_cycles=self.n_cycles+1
self.E += self.energy()
self.E2 += self.energy()**2
self.M += self.magnetisation()
self.M2 += self.magnetisation()**2
return energy,magnetisation
def statistics(self):
Eav = self.E/self.n_cycles
E2av = self.E2/self.n_cycles
Mav = self.M/self.n_cycles
M2av = self.M2/self.n_cycles
# complete this function so that it calculates the correct values for the averages of E, E*E (E2), M, M*M (M2), and returns them
return Eav,E2av,Mav,M2av
Task 5

Introduction to Monte Carlo simulation
Task 6
So the time it takes is:
It would take years to get a single value of .
Task 8
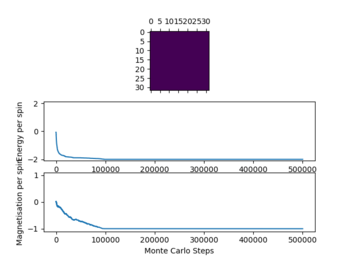
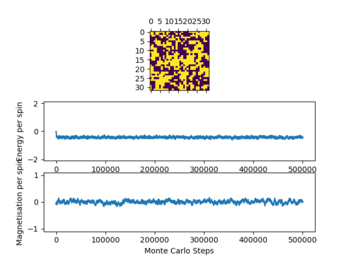
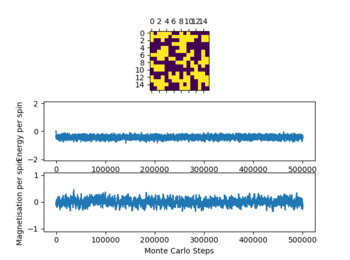
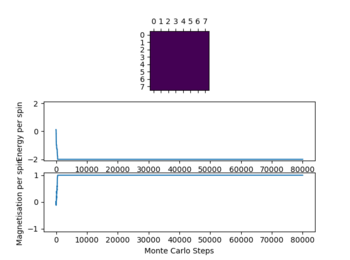
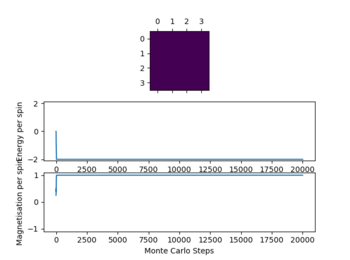
Below the thermal energy isn't great enough to flip spins readily, therefore the lowest energy state dominates over the entropy. If the temperature increases above the curie temperature the entropy dominates and no spontaneous magnetization occurs. Therefore we expect
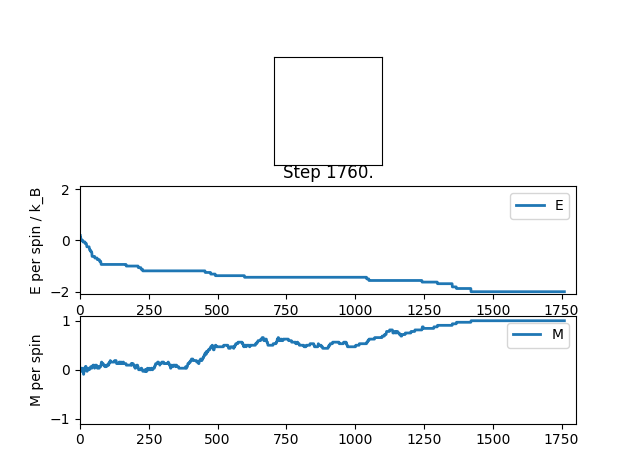
output of statistics file: Averaged quantities:
E = -1.47240896359
E*E = 2.32102153361
M = 0.569695378151
M*M = 0.43897551208
In the figure 2 the system reaches equilibrium at the lowest energy state as all spins align, therefore the system is at the lowest energy. Due to the small temperature the entropy doesn't dominate or contribute.
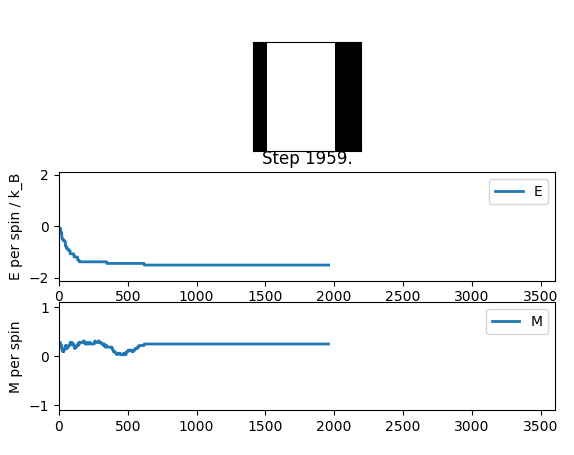
output of statistics file:
Averaged quantities:
E = -1.43791592724
E*E = 2.10344283773
M = 0.233543145519
M*M = 0.0570023950477
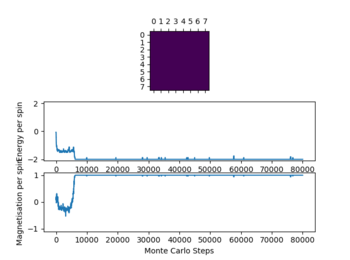
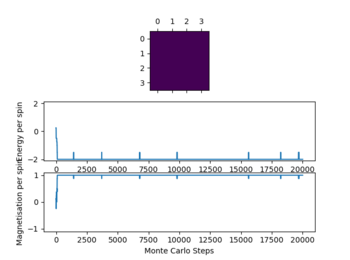
Even thou the system doesn't change in energy anymore it hasn't reached equilibrium, this is due to the fact that it is in the metastable region, where the simulation is at a local minimum and therefore doesn't change anymore as not enough energy is available to flip spins enough to overcome the energy barrier to the global minimum.
Accelerating the code
Task 10
Code of the accelerated function:
import numpy as np
class IsingLattice:
E = 0.0
E2 = 0.0
M = 0.0
M2 = 0.0
n_cycles = 0
def __init__(self, n_rows, n_cols):
self.n_rows = n_rows
self.n_cols = n_cols
self.lattice = np.random.choice([-1,1], size=(n_rows, n_cols))
def energy(self):
"Return the total energy of the current lattice configuration."
colums1=np.multiply(self.lattice,np.roll(self.lattice,1,axis=1))
rows1=np.multiply(self.lattice,np.roll(self.lattice,1,axis=0))
sumpart=np.sum(colums1)+np.sum(rows1)
energy = (-1)*sumpart
return energy
def magnetisation(self):
"Return the total magnetisation of the current lattice configuration."
magnetisation = np.sum(self.lattice)
return magnetisation
def montecarlostep(self, T):
# complete this function so that it performs a single Monte Carlo step
energy = self.energy()
#the following two lines will select the coordinates of the random spin for you
random_i = np.random.choice(range(0, self.n_rows))
random_j = np.random.choice(range(0, self.n_cols))
#the following line will choose a random number in the rang e[0,1) for you
random_number = np.random.random()
#change spin of random number
self.lattice[random_i,random_j] = -self.lattice[random_i,random_j]
#find delta E
newe=self.energy()
deltae=newe-energy
#determine whether new values can be accepted straigth away, no else function needed as the values are already changed
if deltae > 0:
expocalc=np.exp(-deltae/T)
if random_number>expocalc:
#change lattice back as it below R
self.lattice[random_i,random_j] = -self.lattice[random_i,random_j]
energy=self.energy()
magnetisation=self.magnetisation()
#summations of values
self.n_cycles=self.n_cycles+1
if self.n_cycles >0:
self.E += self.energy()
self.E2 += self.energy()**2
self.M += self.magnetisation()
self.M2 += self.magnetisation()**2
return energy,magnetisation
def statistics(self):
Eav = self.E/(self.n_cycles-0)
E2av = self.E2/(self.n_cycles-0)
Mav = self.M/(self.n_cycles-0)
M2av = self.M2/(self.n_cycles-0)
cycles=self.n_cycles
# complete this function so that it calculates the correct values for the averages of E, E*E (E2), M, M*M (M2), and returns them
return Eav,E2av,Mav,M2av,cycles
Task 9 and Task 11
Before accelerating the simulation to run 2000 Monte Carlo steps took 14.90211159 seconds on average of 8 repeats with a standard deviation of 0.10146431 seconds. After acceleration it takes on an average of 8 repeats 0.449982787 seconds to run 2000 Monte Carlo steps with a standard deviation of 0.008642247 seconds.
The effect of temperature
Task 12 and Task 14
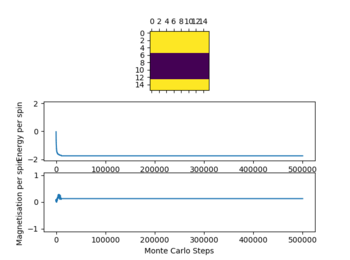
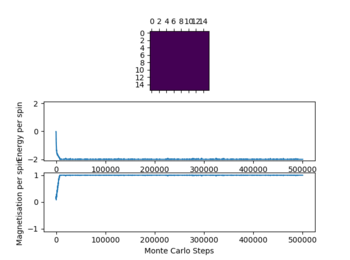
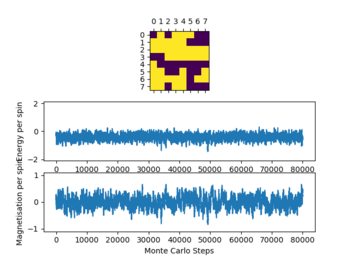
For all lattice sizes timesteps at T=0.25, T= 1 and T= 5 were recorded to show the equilibrium structure at these points.
In the table (Figure 4) above the ILfinalframe graphs are shown at the timestep chosen alongside the temperatures the simulations were run.
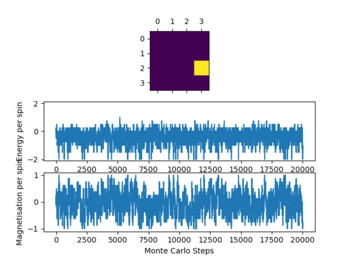
Due to the data obtained a number of 80000 steps was chosen ignoring the first 20000 steps. This was done as with increasing temperature (while ) the simulation needs more time to reach equilibrium as seen above. This effect becomes more pronounced with increasing T. This occurs due to the increased influence of the entropy upon the energy as it is T dependent, which give a lot of local minima when approaching the Curie temperature. This effect can especially be observed in the graphs of the 32x32 lattice.
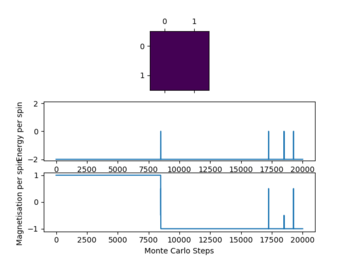
Not all simulations reach the global minimum especially at lower temperatures (e.g 16x16 lattice at T=0.25), this occurs as the simulations reaches a local energy minimum with a thermal energy that is too low to overcome an energy barrier between this local minimum and the global minimum. For all lattice sizes the averages were only recorded once the system reached equilibrium, to ensure that this is the case also for higher temperatures above 1 the steps ignored were significantly larger than the values seen when the reaction reaches equilibrium.
The graphs in figure 4 also show the significant effect of lattice size upon the magnetization and energy values changes. For the 2x2 lattice a change in flip can cause the system to flip magnetization as seen in one of the graphs. Note that the magnitude of the magnetization is important no the sign. Once the magnetization reaches its maximum magnitude the system is at equilibrium as all spins are aligned which gives the biggest magnetic moment.
Task 13 and task 14
| Lattice | Error bars of the mean Variance in Energy at equilibrium | Error bars of the repeats |
|---|---|---|
| 32x32 |  |

|
| 16x16 |  |

|
| 8x8 |  |

|
| 4x4 | 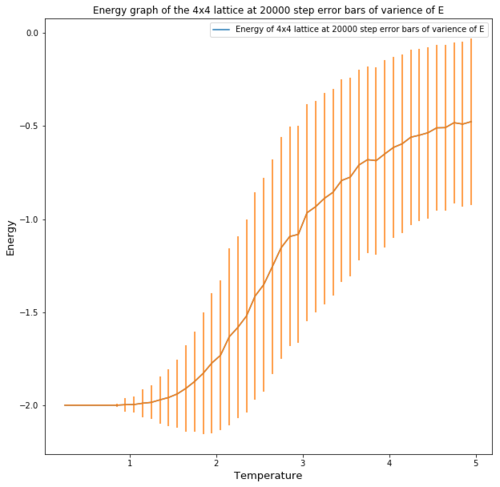 |

|
| 2x2 | 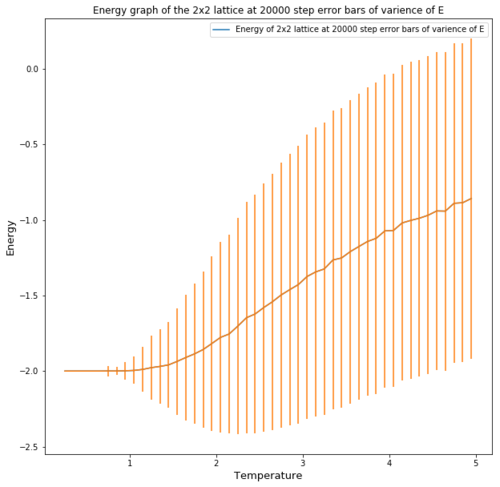 |

|
| Lattice | Error bars of the mean Variance in Magnetization at equilibrium | Error bars of the repeats |
|---|---|---|
| 32x32 |  |

|
| 16x16 |  |

|
| 8x8 |  |

|
| 4x4 |  |

|
| 2x2 |  |

|
Figures 4 and 5 show the effect of the lattice size on the energy per spin and the accuracy of the repeats. One can see that for the energy the error obtained for the energy value at equilibrium is much greater the greater the temperature. This occurs as a higher thermal energy leads to more possible energy states as entropy place a big factor in the energy. The error also decreases significantly with increasing lattice size, this is due to the fact that for a small lattice, flipping one spin causes a large fluctuation in energy, therefore the error is very pronounced, especially at high temperatures where spins flip constantly.
The energy graphs also show that increasing the lattice size makes the phase transition more pronounced and more function like.
Determining the heat capacity
Task 15
with
Thererfore
as
Therefore:
Task 16
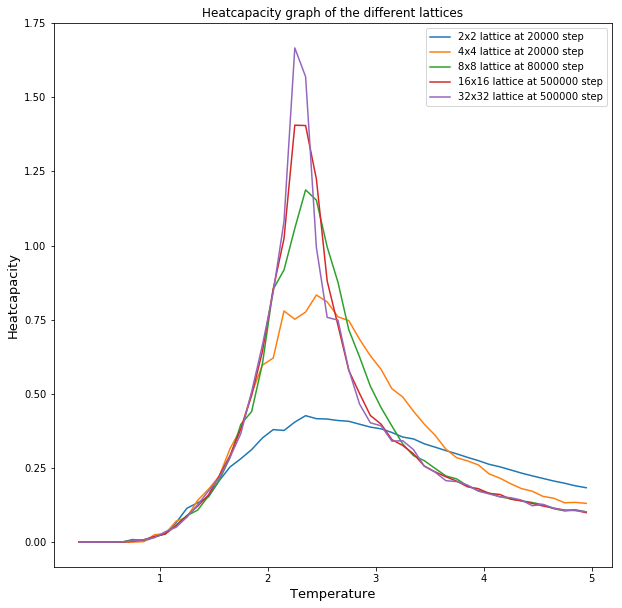
In figure 6 one can see all of the heatcapacities of the different Lattice sizes simulated using the code above. It can be seen that the peak of shifts left and increases with increasing lattice size. The heatcapacity should diverge at the curie temperature, however because we don't use an infinite lattice our heatcapacities don't diverge but reach a peak.
Locating the Curie temperature
Task 17

In figure 7 one can see the difference in the heatcapacity of the C++ data against the one simulated in this experiment. It can be seen that the data is very different around the peak, which suggests that around it the temperature step chosen in the simulation(0.1) was to great in that region. The rest of the plot fits with the C++ data. In future experiments one should decrease the temperature step around the curie temperature to ensure a better fit.
Task 18
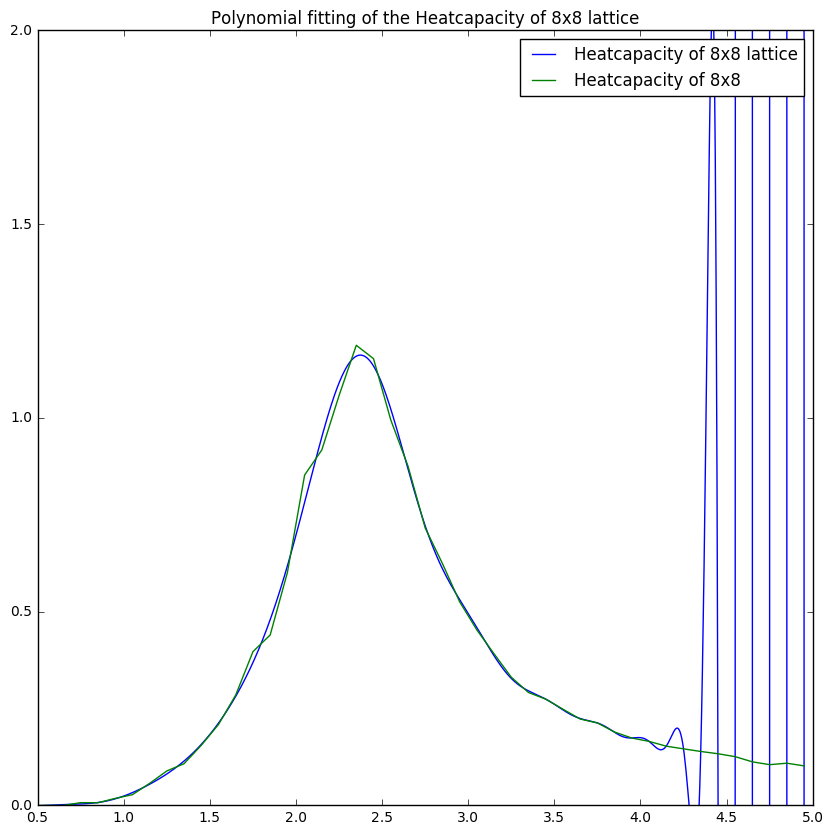
Task 19
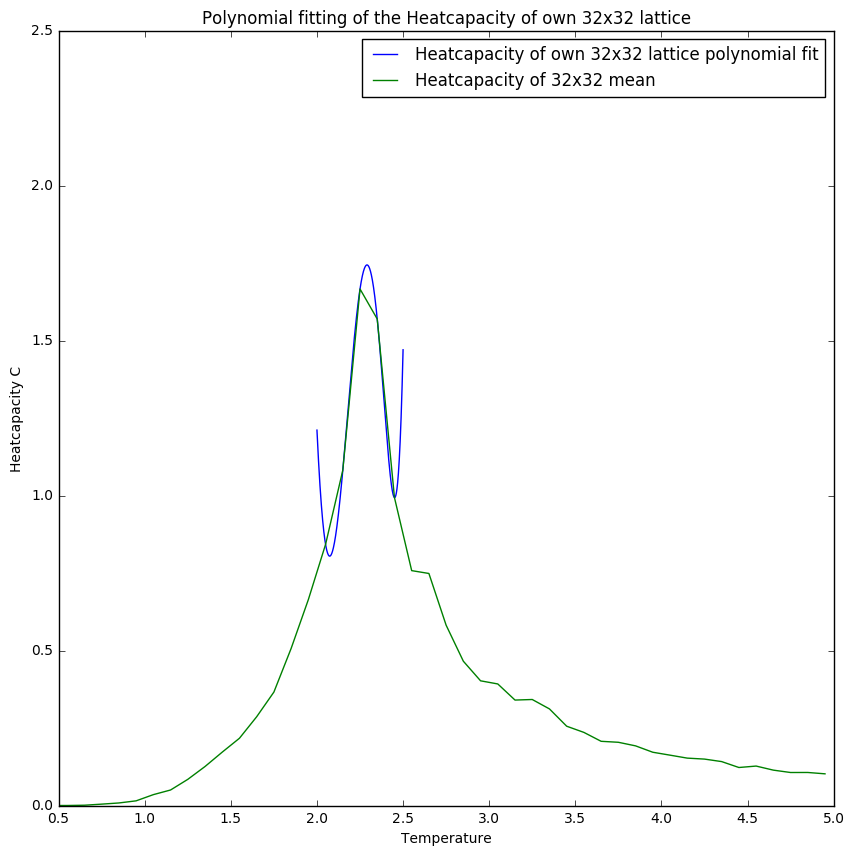
Comparing figures 8 and 9 one can see the importance of selecting the right range for fits as it gives a more reliable fit. This is especially illustrated in figure 10 where the polyfit is much more like the plot obtained from the C++ data.

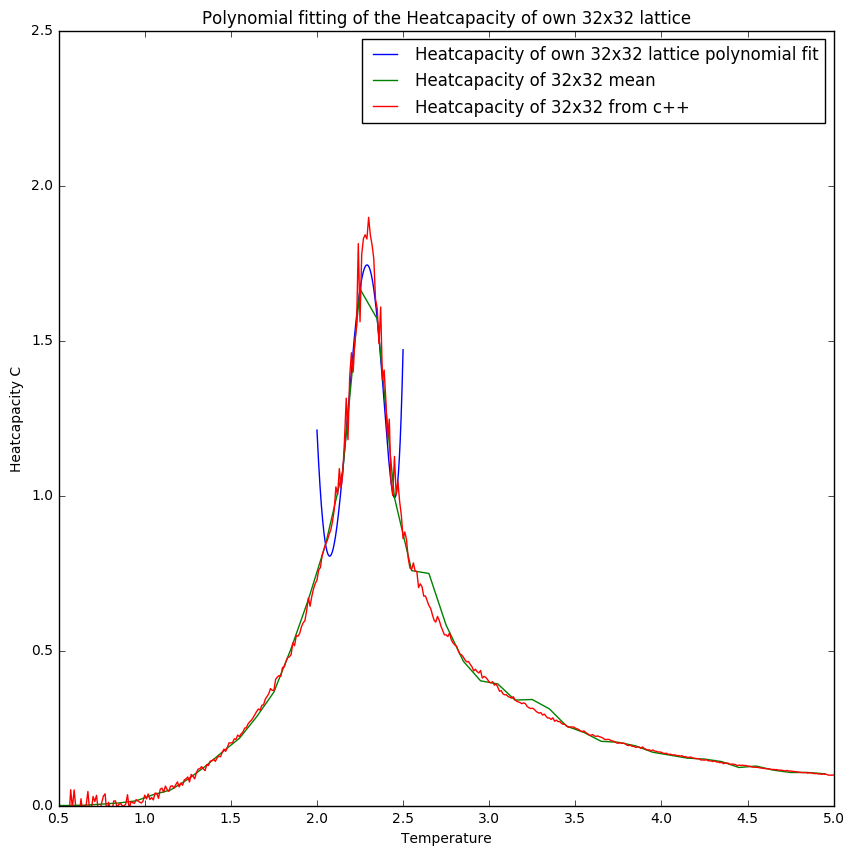
Figures 10 and 11 show that with increasing lattice size the simulation becomes less accurate for the heatcapacity compared to the C++ plots. This could be due to the fact that the heatcapacity at higher lattice sizes becomes more like a function which means that the a small temperature step becomes more significant.
Task 20
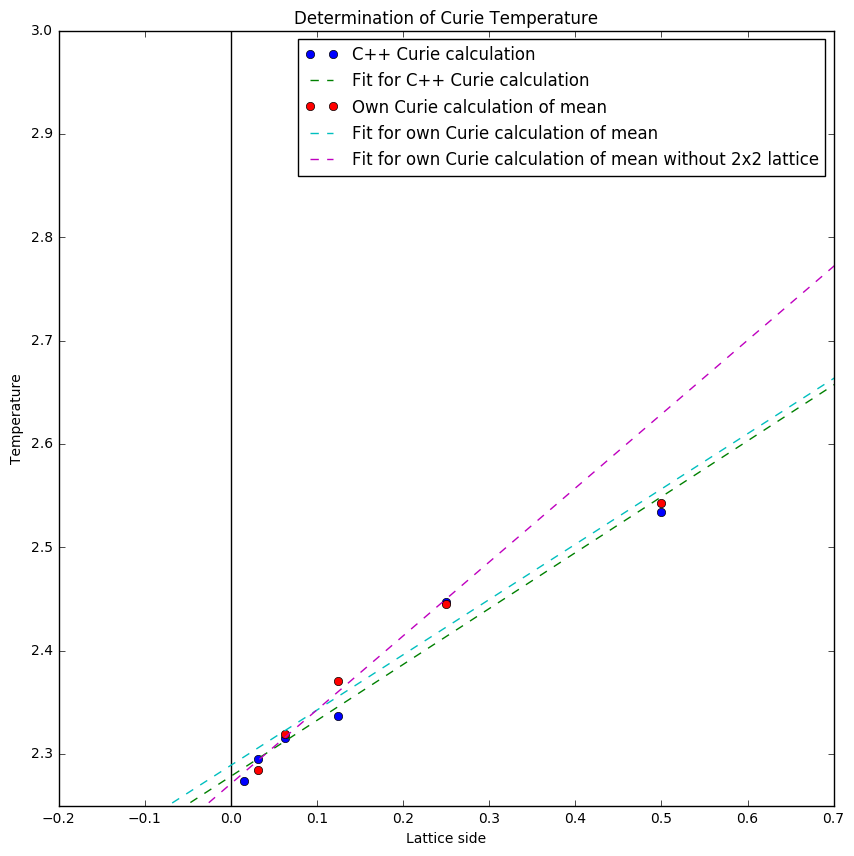
Figure 12 shows the curie temperatures obtained when plotting peak of the heatcapacity against 1 over the lattice size. The intercepts at 0 give the respective curie temperatures:
Own Curie temperature calculation:2.28887220375
C++ Curie temperature calculation: 2.2782708112437811
Selected range of own Curie temperature calculation: 2.2710319004347821
literature value: 2.269[1] (Note the literature value is the exact solution to the 2D IsingLattice model calculated by Onsager)
One can see that the C++ data is more accurate than the simulations done in this experiment. However once the 2x2 lattice heatcapacity was excluded the value of the Curie Temperature became much more accurate. This could be due to the large error associated to the 2x2 Lattice heatcapacity as can be seen in figure 13.
Considering how close the value of the simulation is to the actual value shows the effectiveness of this simulation. The biggest source of error is the limited lattice sizes and the temperature step being to great in the critical region.
| Lattice | Error bars of the heatcapacity of repeats at equilibrium |
|---|---|
| 32x32 | 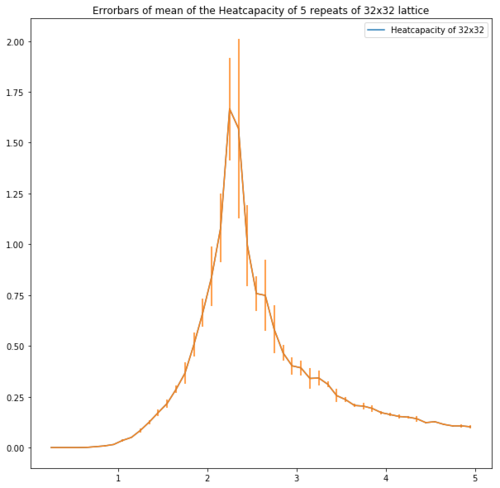
|
| 16x16 | 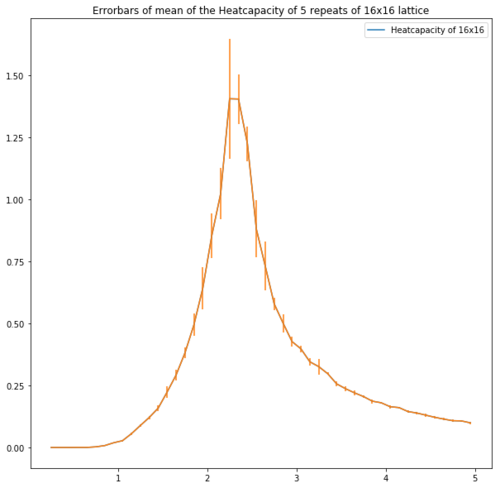
|
| 8x8 | 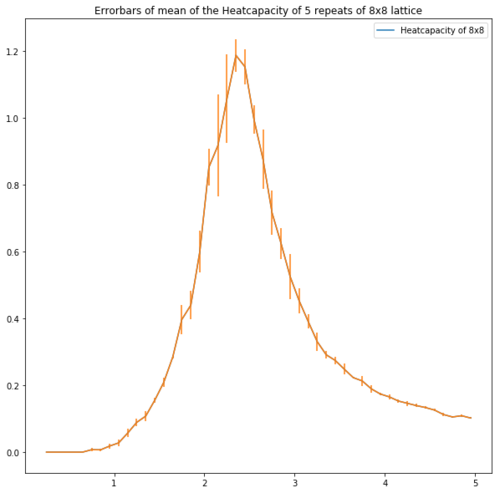
|
| 4x4 | 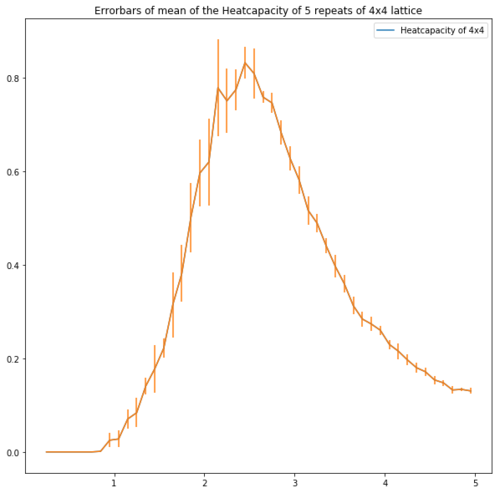
|
| 2x2 | 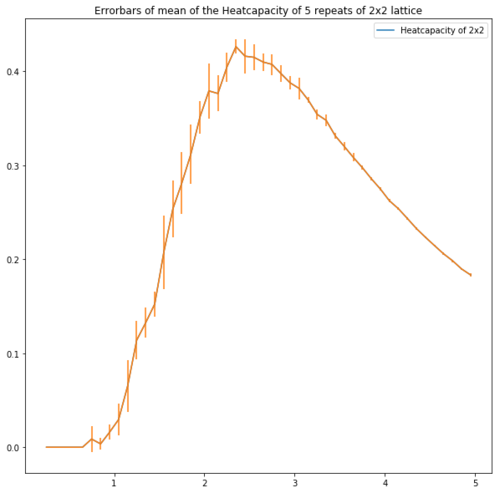
|
Figure 13 shows the error bars of the heatcapacities. These show that for the bigger lattices the error is high around the peak which supports the idea of repeating with a smaller temperature step.
Functions for plotting
Task 13 and 14
def make_plotenergy(dataname,Title,size):
T=dataname[:,0]
E=dataname[:,1]
E2=dataname[:,2]
Eps=E/(size**2)
Eps2=E2/(size**4)
VarE=(Eps2-(Eps)**2)
#sterrorb=(VarE/steps)**0.5
sterrorb=VarE**0.5
figsize(10,10)
title("Energy graph of the "+Title)
xlabel("Temperature",fontsize=13)
ylabel("Energy",fontsize=13)
plot(T,E/(size**2),label="Energy of "+Title)
errorbar(T, E/(size**2), xerr=0, yerr=sterrorb)
legend()
return
def make_plotenergydifferr(dataname,dataerror,Title,size):
T=dataname[:,0]
E=dataname[:,1]
E2=dataname[:,2]
Eps=E/(size**2)
Eps2=E2/(size**4)
sterrorb=dataerror
figsize(10,10)
title("Energy graph of the "+Title)
xlabel("Temperature",fontsize=13)
ylabel("Energy",fontsize=13)
plot(T,E/(size**2),label="Energy of "+Title)
errorbar(T, E/(size**2), xerr=0, yerr=sterrorb)
legend()
return
def make_plotmag(dataname,Title):
T=dataname[:,0]
M=dataname[:,3]
M2=dataname[:,4]
VarM=(M2-(M)**2)
sterrorb=(VarM)**0.5
figsize(10,10)
title("Magnetisation graph of the "+Title)
xlabel("Temperature",fontsize=13)
ylabel("Magnetisation",fontsize=13)
plot(T,M,label="Magnetisation of "+Title)
errorbar(T, M, xerr=0, yerr=sterrorb)
legend()
return
def make_plotmagdifferr(dataname,dataerror,Title,size):
T=dataname[:,0]
M=dataname[:,3]
M2=dataname[:,4]
Mps=M/(size**2)
Mps2=M2/(size**4)
sterrorb=dataerror
figsize(10,10)
title("Magnetisation graph of the "+Title)
xlabel("Temperature",fontsize=13)
ylabel("Magnetisation",fontsize=13)
plot(T,M/(size**2),label="Energy of "+Title)
errorbar(T, M/(size**2), xerr=0, yerr=sterrorb)
legend()
return
def support_plotenergy(dataname,Title):
T=dataname[:,0]
E=dataname[:,1]
VarE=(E2-(E)**2)
#sterrorb=(VarE/steps)**0.5
sterrorb=VarE**0.5
figsize(10,10)
plot(T,E,label="Energy of "+Title)
errorbar(T, E, xerr=0, yerr=sterrorb)
legend()
return
def support_plotenergywithouteer(dataname,Title,size):
T=dataname[:,0]
E=dataname[:,1]
figsize(10,10)
plot(T,E/(size**2),label="Energy of "+Title)
legend()
return
def support_plotmag(dataname,Title):
T=dataname[:,0]
M=dataname[:,3]
M2=dataname[:,4]
VarM=(M2-(M)**2)
sterrorb=(VarM)**0.5
figsize(10,10)
plot(T,M,label="Magnetisation of "+Title)
errorbar(T, M, xerr=0, yerr=sterrorb)
legend()
return
Task 16
def make_plotheatcap(dataname,Title,size):
T=dataname[:,0]
E=dataname[:,1]
E2=dataname[:,2]
heatcap=(E2-(E)**2)/((size**2)*(T**2))
figsize(10,10)
title("Heatcapacity graph of the different lattices")
xlabel("Temperature",fontsize=13)
ylabel("Heatcapacity",fontsize=13)
plot(T,heatcap,label=Title)
#legend()
return
def support_plotheatcap(dataname,Title,size):
T=dataname[:,0]
E=dataname[:,1]
E2=dataname[:,2]
heatcap=(E2-(E)**2)/((size**2)*(T**2))
figsize(10,10)
plot(T,heatcap,label=Title)
legend()
return
Task 17
def support_plotheatcapc(dataname,Title):
T=dataname[:,0]
E=dataname[:,1]
E2=dataname[:,2]
C=dataname[:,5]
k=1.38*10**(-23)
heatcap=(E2-(E)**2)/((T**2))
figsize(10,10)
plot(T,C,label="Heatcapacity of "+Title)
legend()
return
Task 18 and 19
def make_plotheatcapofC(dataname,deg,Title,region1,region2,xlimits,ylimits):
T = dataname[:,0] #get the first column
C = dataname[:,5] # get the second column
#first we fit the polynomial to the data
Tmin = region1 #for example
Tmax = region2 #for example
selection = np.logical_and(T > Tmin, T < Tmax) #choose only those rows where both conditions are true
peak_T_values = T[selection]
peak_C_values = C[selection]
fit = np.polyfit(peak_T_values, peak_C_values, deg) # fit a third order polynomial
#now we generate interpolated values of the fitted polynomial over the range of our function
T_range = np.linspace(Tmin, Tmax, 1000) #generate 1000 evenly spaced points between T_min and T_max
fitted_C_values = np.polyval(fit, T_range) # use the fit object to generate the corresponding values of C
figsize(10,10)
ylabel("Heatcapacity C")
xlabel("Temperature")
title("Polynomial fitting of the Heatcapacity of "+Title+" lattice")
xlim(0.5,xlimits)
ylim(0,ylimits)
plot(T_range,fitted_C_values,label="Heatcapacity of "+Title+" lattice polynomial fit")
Cmax = np.max(fitted_C_values)
Tmax = T_range[fitted_C_values == Cmax]
return Cmax,Tmax
def make_plotheatcapofown(dataname,size,deg,Title,region1,region2,xlimits,ylimits,range1,range2):
T = dataname[:,0] #get the first column
E=dataname[:,1]
E2=dataname[:,2]
#first we fit the polynomial to the data
C=(E2-(E)**2)/((size**2)*(T**2))
Tmin = region1 #for example
Tmax = region2 #for example
selection = np.logical_and(T > Tmin, T < Tmax) #choose only those rows where both conditions are true
peak_T_values = T[selection]
peak_C_values = C[selection]
fit = np.polyfit(peak_T_values, peak_C_values, deg) # fit a third order polynomial
#now we generate interpolated values of the fitted polynomial over the range of our function
T_range = np.linspace(Tmin, Tmax, 1000) #generate 1000 evenly spaced points between T_min and T_max
fitted_C_values = np.polyval(fit, T_range) # use the fit object to generate the corresponding values of C
figsize(10,10)
ylabel("Heatcapacity C")
xlabel("Temperature")
title("Polynomial fitting of the Heatcapacity of "+Title+" lattice")
xlim(0.5,xlimits)
ylim(0,ylimits)
plot(T_range,fitted_C_values,label="Heatcapacity of "+Title+" lattice polynomial fit")
Cmax = np.max(fitted_C_values[range1:range2])
Tmax = T_range[fitted_C_values == Cmax]
return Cmax,Tmax
Task 20
figsize(10,10)
ylim(2.25,3)
xlim(-0.2,0.7)
owncuriefit=polyfit(owncurieviagrad[:,0], owncurieviagrad[:,1],1)
precalccuriefit=polyfit(precalccurieviagrad[:,0], precalccurieviagrad[:,1],1)
owncuriefitofmeansel=polyfit(owncurieviagradofmean[1:,0], owncurieviagradofmean[1:,1],1)
owncuriefitofmean=polyfit(owncurieviagradofmean[:,0], owncurieviagradofmean[:,1],1)
x=arange(-1,5,0.01)
title("Determination of Curie Temperature")
xlabel("Lattice side")
ylabel("Temperature")
plot(x*0,x,linestyle="-",color="black")
#plot(owncurieviagrad[:,0],owncurieviagrad[:,1],marker="o",linestyle="",label="Own Curie calculation")
plot(precalccurieviagrad[:,0],precalccurieviagrad[:,1],marker="o",linestyle="",label="C++ Curie calculation")
#plot(x,x*owncuriefit[0]+owncuriefit[1],linestyle="--",label="Fit for own Curie calculation")
plot(x,x*precalccuriefit[0]+precalccuriefit[1],linestyle="--",label="Fit for C++ Curie calculation")
plot(owncurieviagradofmean[:,0],owncurieviagradofmean[:,1],marker="o",linestyle="",label="Own Curie calculation of mean")
plot(x,x*owncuriefitofmean[0]+owncuriefitofmean[1],linestyle="--",label="Fit for own Curie calculation of mean")
#plot(owncurieviagradofmean[0:-1,0],owncurieviagradofmean[0:-1,1],marker="o",linestyle="",label="Own Curie calculation of mean without 2x2 lattice")
plot(x,x*owncuriefitofmeansel[0]+owncuriefitofmeansel[1],linestyle="--",label="Fit for own Curie calculation of mean without 2x2 lattice")
legend()
references
<references> [1]