Rdm:xray-data
Publishing raw Crystal Data
Crystal structure data comes in broadly four types:
- The "raw" data that is collected by the instrument, comprising diffraction images and from which the space group is determined.
- Semi-processed data, containing so called structure factors, and which is a sub-set of the raw data.
- Publication data, in so called CIF (crystallographic information format) containing the coordinates of the atoms.
- Data in the form of an image representation of the molecule, which can serve highlight any interesting chemical features.
For a full replication of the crystal structure solution, the first type is essential. If the assigned space group of the molecule is accepted as being correct, then the structure factor data can be used to refine the data into a proposed structure for the molecule. Such replication is not possible for the third type (the CIF) file, which can however be used to obtain various structural and other properties of the molecule and for which a CheckCIF validation of this data can be performed. The image of the molecule is useful to humans but less so for machines accessing the data.
Traditional publication of crystal structure data normally involves just the CIF file, possibly a CheckCIF validation and a picture of the molecule. Increasingly however the trend is towards submission of full (raw) instrumental data, which would in principle allow anyone to check the analysis of the data. The CSD (Cambridge structure database) is the normal repository for crystal data. Up to around ten years ago, they only accepted the second two categories, but then started accepting the structure factor data as well. To this date however they have no mechanism for depositing the raw data and this is where the local data repository comes in.
For an example of raw instrument data associated with a research publication, go to DOI:10.14469/hpc/2297 . This is a collection of seven such sets of data, one example of which can be inspected here: DOI:10.14469/hpc/2298 . You will see that it contains a link to the published article DOI:10.1021/acsomega.7b00482 as well as the CIF data deposited at CSD. In turn, the CSD entry DOI:10.5517/ccdc.csd.cc1n9pn9 contains a link back to the raw data for each of the seven structures.
Files needed for publishing raw crystal data
The raw crystal data (a collection of diffractometer images) can be archived into a folder using common compression formats. Three are discussed here. The first is 7-zip, which provides the most efficient compression. If the older zip compression/archive format is used, it should be further compressed using gzip to form a .gzip archive to avoid confusion with a similar process used for archiving raw NMR data files. The general problem of how to tell what is inside a compressed file remains a challenge! In this case, identification can be helped by also uploading a CIF file to in effect identify the data as being crystallographic.
Live Preview
One other file needs explanation. This is an index.html file, which is used to produce a simple visual preview of the molecule. If desired, this could be modified to produce a dynamic interactive 3D model of the molecule using the code JSmol (not illustrated for this deposition, but easily added). Details on this operation will shortly be added.
Adding the raw data citation to the CSD deposition
This is done in the enhance data option of the deposition process using https://www.ccdc.cam.ac.uk/deposit/ via associated DOIs

The above shows the sequence for uploading data to CSD.
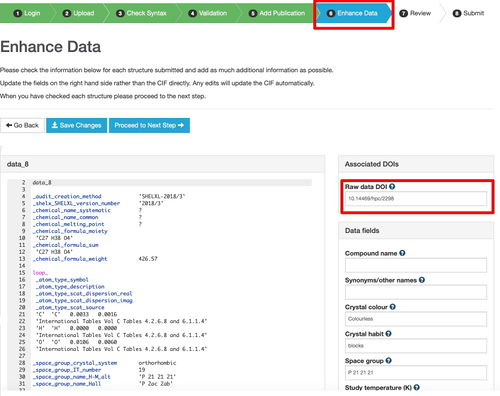
In turn, when the article citing the data is published, along with the CSD entry, the DOIs for both these sources can be added to the record on the raw data repository, thus completing fully bidirectional links from each data source to the other. The metadata records for each deposition now contain reference to the other.
