Rdm:intro
Research Data Management in Chemistry (RDM)
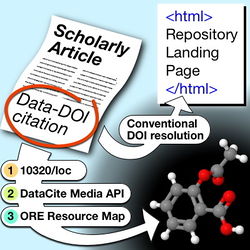
A set of hints and procedures for RDM in chemistry. Because research data in chemistry covers a very wide field, this is not meant to be an inclusive or even remotely complete set of instructions. It documents actions that can be currently done (or almost done) rather than just aspirations for the future. If you know of any resource that should be listed here, please add them yourself.
General background information
- Flowchart for Researchers to meet Imperial College London RDM policy
- More detailed report with use case examples
- Computational chemistry use cases
Why a Data Repository?
Why use a data repository rather than SI (supporting information) submitted to a journal? Here are some reasons.
- SI often takes the form of a single monolithic document which can balloon to 200 pages or more. It does not conform to FAIR (findable, accessible, interoperable and re-usable) Data, and is held in a format (PDF) that was not designed to hold data.
- A data repository is (or can be) FAIR:
- It is findable (it can be searched, i.e. indexed by not only by Google but by data-savvy sites such as Datacite.org) and these searches are strongly guided by metadata.
- Data repositories mandate that metadata be created to go with the data, thus facilitating FAIR. Creating, handling and exposing metadata takes a specialised environment, one simply not provided by SI in PDF form on a publisher's website.
- It is accessible via its own DOI (digital object identifier)
- The data in a repository is required to conform to standard types appropriate for its content. There are many such interoperable types; for example JCAMP-DX for spectroscopy, Molfile for atom coordinates, CML (chemical markup language) or more specialised types designed for specific types of program such as e.g. MestreNova or Gaussview.
- Re-usable means the data can be quickly and easily read by standard analysis programs to allow its re-use in perhaps a context different from the one it was created for.
- Retrieval of such data from a repository can be automated by software using the assigned DOI and need not require eg processing by a human. If need be it can be usefully mined to increase its value. Examples are listed below.
Working examples of RDM in action
- Log into https://portal.hpc.imperial.ac.uk/, or the UPORTAL as referred to below. This environment is designed for security and to hold some types of metadata associated with you the researcher. To access, you need appropriate credentials (normally your College email login and password).
- Personal metadata can be set up once to include
- Your ORCID identifer, a now standard method for disambiguating yourself from others with the same or similar name.
- Your desired embargo policy (more later).
- Where you want to publish your data; Figshare is one of three repositories currently available.To use Figshare, you should click on the link, when you will be redirected to the Figshare site. If you are a new user to Figshare, you should create yourself an account there. With an account you will then be asked to allow UPORTAL to read and write data to Figshare. Click on allow.
- Back at the UPORTAL you should now see that Figshare credentials are present.
- Invoke the uploader. This enables the creation of a basic set of metadata (including your ORCID), along with broad topics in chemistry.
- A basic description field could contain e.g. the relevant notes from the experimental section of a thesis.
- It is assumed (but is not mandatory) that your data will relate to a specific molecule and you can create specific metadata such as an InChI identifier. This is done by adding a file containing a Chemdraw .cdx file with the structure of your molecule. This is then converted to an InChI identifier by the UPORTAL.
- Other files can be added, eg Spectral PDF, or ideally JCAMP-DX files of the spectrum.
- Uploading will create a PRIVATE fileset on Figshare, where you can check the details and add additional data such as co-authors, or links (DOIs) to the forthcoming journal article to which the data might relate to.
- At this stage, Figshare assigns a DOI to the data, which can be quoted in any article in which the data is referred to
- Links to the authors' accepted version deposited into the institutional repository SPIRAL can also be added and to any other relevant materials.
- You would also need to action this file set on the Figshare site to release it to public mode at the appropriate moment. At this point, you can also generate a ShortDOI for the data.
- The UPORTAL will keep a record of your deposition, with metadata such as the deposition date, and the Figshare DOI assigned to it. UPORTAL allows filtered searches to track down earlier depositions, and you can also release the deposition to public mode from here rather than Figshare.
- Examples of the outcome of this process
- DOI:10.6084/m9.figshare.777773 or shortDOI DOI:rnf illustrates spectral data, and where you can also find a link to the published article associated with the data: DOI:10.1021/jo401316a .
- DOI:10.6084/m9.figshare.1342036 , shortDOI DOI:2zb includes some computer scripts/programs associated with a publication.
- DOI:10.6084/m9.figshare.988346 , shortDOI DOI:tb3 includes a host of supporting information (including Wiki source code documentation) relating to a laboratory experiment, serving the purpose of helping others introduce that experiment into their own laboratory.
Figshare provide a stand alone tool for uploading data. It is configured with the user's Figshare account credentials, and thereafter it is simply a drag-n-drop operation to upload the files. This process involves no metadata capture, and this has to be added manually using the interface on the Figshare site. The result is private until made public. This represents an alternative way of creating the depositions shown in example 1 above.
Data files on their own require some action by their acquirer to become useful, such as using a graphical program to eg display 3D coordinates for a molecule, or spectra, etc. Some of this wrapping can be done beforehand, and the result uploaded to a data repository in more immediately visual/usable form. This requires more work by the depositor, and the process will not be described here. A few examples suffice to illustrate:
- DOI:10.6084/m9.figshare.1299202 , shortDOI: DOI:4fw relates to data for a study of the mechanism of the bromination of benzene, and is presented in the form of an interactive table whereby the required data is retrieved (using its DOI) from a repository and presented to the user in rotating, vibrating interactive form. The table itself contains reference to multiple datasets if the reader wishes to access them for downloading rather than visual inspections.
- DOI:10.6084/m9.figshare.1115056 , shortDOI: DOI:ttf takes the form of a clickable figure, where regions of the diagram respond by producing two new browser TABS, one showing in interactive visual form a presentation of the data and the other TAB showing the data repository source of that data. The user can decide which of the two tabs to use.
- DOI:10.6084/m9.figshare.1266197 shortDOI:DOI:xn3 is an advanced example containing several layers of RDM.
- The document takes the form of a HTML page acting as the table of contents for data.
- This HTML replaces the normal landing page when the DOI is resolved (this can be done by Figshare on request).
- The HTML contains scripts that in turn directly invoke data from a repository and then display it on the HTML Web page.
- This mechanism means that the reader need not handle the data from a repository themselves; they merely see a visual interactive presentation of that data.
Deposition of Computational modelling data using UPORTAL
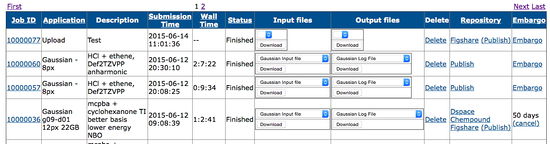
This is a more specialised use for the UPORTAL tool mentioned in example 1. It acts to simplify the submission of computational molecular models to a high performance (HPC) resource, to return the model already annotated with appropriate metadata and to keep a record of all such work. The depositions illustrated in example 3 above were generated using this method.
- The concept is of a workflow, or sequence of actions.
- The user selects an application they wish to use on the HPC and a queue appropriate for the resources it is expected to need.
- A short description is added to remind the user, to help in future searches, and also as metadata for any future deposition into a data repository
- The system records the submission date and times and the wall time, the elapsed time actually taken.
- The input files needed to define the problem, together with output files are then listed as available for downloading and imspection
- A delete button allows couple deletion of a (presumably failed or otherwise useless)entry.
- A repository section allows publication to a specified repository (as defined in the user's profile) having the publication DOI returned to the UPORTAL.
- With Figshare the publication remains private until the PUBLISH option is selected to make it public.
- With DSpace, the publication is made public immediately.
- Finally, the user can choose to EMBARGO the item. This keeps the data private but will send the user an email after the embargo period is reached as a reminder that further action is needed. The embargo can then be extended if wished, or the data published, following appropriate links embedded in the email.
An example of the outcome of this workflow is DOI:10.14469/ch/191324 , where you can see the metadata gathered for the deposit and the various files in the collection. This has the advantage that all the metadata associated with the deposition becomes available to search at DataCite, a feature that is unique to a data repository and is not available with conventional supporting information.
The Chemspider synthetic deposition pages
This is an example of a publisher (the Royal Society of Chemistry) offering a database-repository which allows experimental data to be archived and assigned a DOI, DOI:10.1039/SP501 .
The CCDC CIF Deposition service
This page can be used to deposit a crystal structure and receive back an assigned DOI for the data. An example of how this data is exposed can be seen at DOI:10.5517/CC11TJ7M , shortDOI: DOI:5c8
The Zenodo General data deposition service
Zenodo is run by CERN, experts in collecting and deposition large data sets. Whilst it is relatively little used for chemistry (--Rzepa (talk) 08:53, 17 June 2015 (BST)) it does represent a (currently) free service.
- The deposition page requires you to register an account, which you can in fact do using your ORCID credentials (a method which is gratifyingly on the increase). The metadata gathering is relatively generic. One can also specify a community that will be alerted to your data. Currently, the deposition and metadata specification is all manual, but we are investigating how much of this could be automated by using a deposition API.
- Once a deposition is completed, a DOI is assigned, e.g. DOI:10.5281/zenodo.18632 as part of a community
Chemical Wikidata
A newcomer to data deposition is Wikidata. This holds data in more highly declared semantic form, and its prime purpose is to populate Wikipedia pages. It is listed here more as a place holder whilst we learn about its potential.
Examples of RDM cited in chemistry journal articles.
- DOI:10.1021/jo401316a contains an extensive experimental section listing the DOIs of individual calculations, and these are also included in two interactivity boxes in the HTML version of th article, known as WEOs (web-enhanced objects). An example of one such dataset is DOI:10.14469/ch/13672
- DOI:10.1021/ed500398e lists both conventional SI in the acknowledgements and references 17 and 18 to data repository content.
- DOI:10.1002/anie.201407751 lists ref 20 for both computational data as DOI:10.6084/m9.figshare.1115056 , short:DOI:ttf and also crystal data in a separate data repository (the CCDC), with a DOI:10.5517/CC11TJ7M , short:DOI:5c8
- DOI:10.1002/anie.201409672 cites an interactive data table for details of computational data: DOI:10.6084/m6089.figshare.1167503 , short:DOI:vmh
- DOI:10.1039/C3SC53416B gives a dataDOI in the caption for all the tables of numerical information presented, and interactive presentational DOIs for all the figures. One example is included here: DOI:qd8 .
- DOI:10.1186/s13321-015-0081-7 is an article about how to access cited data, and includes ref 27, DOI:10.6084/m9.figshare.1266197 short:DOI:xn3 as a working example.
Background information to activity in Chemical RDM
- Some experiments in RDM conducted as part of a Imperial College funded project in 2014. These include a number of examples of data searches using DataCite.
For more information please contact --Rzepa (talk) 09:06, 15 June 2015 (BST)
