PYP : Photoactive yellow protein
PYP
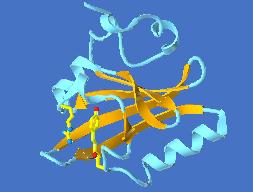
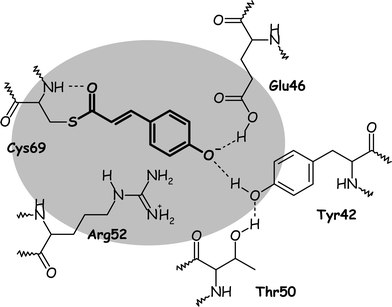
PYP, the Photoactive Yellow Protein, is a small water-soluble protein extracted from the cytosol of the halophilic purple bacterium Halorhodospira halophila. PYP is thought to mediate the phototactic response of the bacterium against blue light. Its chromophore is the deprotonated trans-p-hydroxycinnamic acid covalently linked, via a thioester bond, to the unique cysteine residue of the protein. Upon blue-light irradiation, PYP undergoes a photocycle. As for rhodopsins, the trans to cis isomerization of the chromophore was shown to be the first overall step of this photocycle.
PDB : Protein Data Bank
In order to run some calculations about a protein, you need an input file ! You can find the protein already drawn in the Protein Data Bank (PDB) website. Just search your molecule in this data base which regroup most research already done about protein. Choose a file, and then click on Download files --- > PDB file (Text).
The PDB file has an extension which is .pdb, and this extension can be directly open by GaussView (the short open is not possible, but Open GaussView and then click on File ---> Open and then select your .pdb file).
Break
If the coordinates of the hydrogens are not mentioned in the PDB file, GaussView will add the hydrogens automatically but so not necessary with the right angles and bond length. So be careful.
First calculation
Now you have the molecule that you want to study which is drawn so do exactly what you did usually so as to run a calculation using GaussView.
Just begin to run a single point energy calculation with AMBER.
You should get an error message with your first calculation (if not you are very lucky and so try to run the different calculations that you would like to run).
Normaly you should have so an error message which could be one of the different that are described in the AMBER part.
If it is a missing atom type :
have a look through your input and find the atom which has no atom type, and give it one thanks to the different proposed by the AMBER data base.
If it is missing MM parameters :
in this case operate very carefully as I mentioned below and do not be afraid to do it step by step.
First just consider the Bond length that are missing, identify the atoms corresponding in the molecule (with GaussView), and check for each of them that the atom type assigned by GaussView is the right one ! If the atom typ is wrong, so you have to change it in the input file. Once all the atom type verified (and changed for the wrong one) run again the calculation.
Now you should have some new missing parameters (or not!), if you have some new one have a look at the new atoms implicated in the new bond length missing and check if the atom types assigned are the right, if not change it...
And then run again the calculation...
When the Bond length missing do not involved some atom types not well assigned, then the Bond length and angles still missing have to be defined manually thanks to the AMBER data base (which is a cut of the AMBER paper).
Advice Break In your protein you have a chromophore and using ONIOM enables you to run calculations with a high level of accuracy for this chromophore, so if some bond lengths or angles just involved atoms of the chromophore define them as 0.0
HrmStr1 * * 0.0 0.0 HrmBnd1 * * * 0.0 0.0
You can do this big approximation because/thanks to ONIOM theory : indeed an AMBER calculation will be run on the full protein and on the chromophore in order to deduce the contribution of the chromophore of the energy of the full protein (because concerning the chromophore the energy will be the one given by the high level of theory).
Example : PYP with 3PHY.pdb file
How to run the calculation ?
Download the .pdb file on the Protein Data Bank website : 3phy:PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED), SOLUTION STRUCTURE, NMR, 26 STRUCTURES
And try to run an AMBER calculation
#p amber geom=connectivity
So you should get this error message :
Missing atomic parameters for atom 1914 IAtTyp= 20000000 Missing atomic parameters.
Thus the atom 1914 is not define (atom type not define).
Indeed the O is not defined
O- -9.64963601 -6.71154940 3.64049197
Have a look at the environment : the O is an O type so write
O-O- -9.64963601 -6.71154940 3.64049197
Run again the calculation after this change.
An other error message should appear :
Missing atomic parameters for atom 1923 IAtTyp= 20000000 Missing atomic parameters.
Define this other atom and run again the calculation.
This time : a new error message, all the parameters below are missing, so have a look at the different atoms and atom types assigned by GaussView
Include all MM classes Bondstretch undefined between atoms 40 41 CM-N2 Bondstretch undefined between atoms 42 44 CM-N2 Bondstretch undefined between atoms 43 51 CA-H5 Bondstretch undefined between atoms 1003 1913 S-C Bondstretch undefined between atoms 1610 1611 CM-N2 Bondstretch undefined between atoms 1612 1614 CM-N2 Bondstretch undefined between atoms 1613 1621 CA-H5 Bondstretch undefined between atoms 1780 1782 CM-N2 Bondstretch undefined between atoms 1915 1924 CM-HC Bondstretch undefined between atoms 1916 1925 CM-HC Angle bend undefined between atoms 36 39 40 CT-CT-CM Angle bend undefined between atoms 39 40 41 CT-CM-N2 Angle bend undefined between atoms 40 41 43 CM-N2-CA Angle bend undefined between atoms 40 41 49 CM-N2-H Angle bend undefined between atoms 40 42 44 CM-CM-N2 Angle bend undefined between atoms 41 43 51 N2-CA-H5 Angle bend undefined between atoms 41 40 42 N2-CM-CM Angle bend undefined between atoms 42 44 1930 CM-N2-H Angle bend undefined between atoms 42 44 43 CM-N2-CA Angle bend undefined between atoms 44 42 50 N2-CM-H4 Angle bend undefined between atoms 44 43 51 N2-CA-H5 Angle bend undefined between atoms 649 648 650 O2-C-OH Angle bend undefined between atoms 1002 1003 1913 CT-S-C Angle bend undefined between atoms 1003 1913 1914 S-C-O Angle bend undefined between atoms 1003 1913 1915 S-C-CM Angle bend undefined between atoms 1606 1609 1610 CT-CT-CM Angle bend undefined between atoms 1609 1610 1611 CT-CM-N2 Angle bend undefined between atoms 1610 1611 1613 CM-N2-CA Angle bend undefined between atoms 1610 1611 1619 CM-N2-H Angle bend undefined between atoms 1610 1612 1614 CM-CM-N2 Angle bend undefined between atoms 1611 1613 1621 N2-CA-H5 Angle bend undefined between atoms 1611 1610 1612 N2-CM-CM Angle bend undefined between atoms 1612 1614 1931 CM-N2-H Angle bend undefined between atoms 1612 1614 1613 CM-N2-CA Angle bend undefined between atoms 1614 1612 1620 N2-CM-H4 Angle bend undefined between atoms 1614 1613 1621 N2-CA-H5 Angle bend undefined between atoms 1775 1778 1779 CT-CT-CM Angle bend undefined between atoms 1779 1780 1782 CM-CM-N2 Angle bend undefined between atoms 1779 1781 1784 CM-CM-CM Angle bend undefined between atoms 1780 1782 1783 CM-N2-CA Angle bend undefined between atoms 1780 1782 1793 CM-N2-H Angle bend undefined between atoms 1780 1779 1781 CM-CM-CM Angle bend undefined between atoms 1781 1783 1785 CM-CA-CA Angle bend undefined between atoms 1782 1783 1785 N2-CA-CA Angle bend undefined between atoms 1782 1780 1792 N2-CM-H4 Angle bend undefined between atoms 1784 1786 1787 CM-CA-CA Angle bend undefined between atoms 1784 1786 1796 CM-CA-HA Angle bend undefined between atoms 1913 1915 1924 C-CM-HC Angle bend undefined between atoms 1915 1916 1925 CM-CM-HC Angle bend undefined between atoms 1916 1917 1918 CM-CA-CA Angle bend undefined between atoms 1916 1917 1922 CM-CA-CA Angle bend undefined between atoms 1916 1915 1924 CM-CM-HC Angle bend undefined between atoms 1917 1916 1925 CA-CM-HC Angle bend undefined between atoms 1919 1920 1923 CA-C-O Angle bend undefined between atoms 1921 1920 1923 CA-C-O MM function not complete
Many atoms are not well defined by GaussView, so change them.
Finally you should have change the following atoms
< 40 C-CM < 41 N-N2 < 42 C-CM < 43 C-CA < 44 N-N2 < 649 O-O2 < 650 O-OH < 1610 C-CM < 1611 N-N2 < 1612 C-CM < 1613 C-CA < 1614 N-N2 < 1779 C-CM < 1780 C-CM < 1781 C-CM < 1782 N-N2 < 1783 C-CA < 1784 C-CM
By the following one
> 40 C-CC > 41 N-NA > 42 C-CV > 43 C-CR > 44 N-NB > 649 O-OH > 650 O-O > 1610 C-CC > 1611 N-NA > 1612 C-CV > 1613 C-CR > 1614 N-NB > 1779 C-C* > 1780 C-CW > 1781 C-CB > 1782 N-NA > 1783 C-CN > 1784 C-CA
Now run again the calculation and you have to define manually the other bond lengths and angles missing.
So finally you should have at the end of your input the following MM parameters
HrmStr1 S C 227.0 1.810 HrmStr1 NB H 434.0 1.010 HrmStr1 * * 0.0 0.0 HrmBnd1 CV NB H 30.0 120.0 HrmBnd1 CR NB H 30.0 120.0 HrmBnd1 O2 C OH 80.0 125.0 HrmBnd1 CT S C 62.0 100.0 HrmBnd1 S C O 80.0 124.0 HrmBnd1 S C CM 80.0 125.30 HrmBnd1 * * * 0.0 0.0
Input, output fair
One atom has no atom type : Media:wiki_test_3phy_1.gjf Media:wiki_test_3phy_1.log
One other atom has no atom type : Media:wiki_test_3phy_2.gjf
Media:wiki_test_3phy_2.log
Now we know that we have some MM parameters missing : begin to check the atoms types concerned Media:wiki_test_3phy_3.gjf
Media:wiki_test_3phy_3.log
First changes operated concerning atoms num 40 41 42 43 44 1610 1611 1612 1613 1614 1780 1782, and then run wiki_test_3phy_4
Media:wiki_test_3phy_4.gjf Media:wiki_test_3phy_4.log
Change atom type of atom 1779 and then run wiki_test_3phy_5
Media:wiki_test_3phy_5.gjf Media:wiki_test_3phy_5.log
Change atom type of atom 1781 and run wiki_test_3phy_6
Media:wiki_test_3phy_6.gjf Media:wiki_test_3phy_6.log
Change atom type of atom 1784 and then run wiki_test_3phy_7
Media:wiki_test_3phy_7.gjf Media:wiki_test_3phy_7.log
So this last should be ok
Media:wiki_test_3phy_8.gjf Media:wiki_test_3phy_8.log
Atoms charges
As you can see in order to run an AMBER calculations the charges of all the atoms have to be specified. GaussView is able to specify many charges (the charges correspondent to different amino acid mentioned in the AMBER paper).
N-N3-0.159200 0 22.53936399 -2.55554940 12.12649197 L C-CT-0.022100 0 21.34836399 -2.49054940 11.23349197 L C-C-0.612300 0 20.19836399 -3.28254940 11.85849197 L O-O--0.571300 0 20.40836399 -4.18754940 12.64249197 L C-CT-0.086500 0 21.70036399 -3.08954940 9.87049197 L C-CT-0.033400 0 21.58536399 -2.01054940 8.79149197 L S-S--0.277400 0 19.86636399 -1.89254940 8.23649197 L C-CT--0.034100 0 20.13836399 -0.68754940 6.91449197 L H-H-0.198400 0 22.76336399 -3.55054940 12.33349197 L H-H-0.198400 0 23.35136399 -2.10754940 11.65549197 L H-H-0.198400 0 22.33636399 -2.05454940 13.01449197 L H-HP-0.111600 0 21.04836399 -1.46054940 11.10849197 L H-HC-0.012500 0 22.71136399 -3.46954940 9.89449197 L H-HC-0.012500 0 21.01736399 -3.89554940 9.64449197 L H-H1-0.029200 0 21.89836399 -1.05954940 9.19849197 L H-H1-0.029200 0 22.21736399 -2.26954940 7.95549197 L H-H1-0.059700 0 20.92536399 -1.04054940 6.26349197 L H-H1-0.059700 0 19.22536399 -0.56354940 6.34849197 L H-H1-0.059700 0 20.42636399 0.25845060 7.34249197 L
But some charges could be missing
O-O--0.567900 0 3.95036399 -10.02254940 -0.92050803 L C-CT--0.182500 0 6.36136399 -7.53054940 -0.90950803 L H-H-0.271900 0 6.06136399 -7.38354940 -3.47850803 L H-H1-0.082300 0 6.29536399 -9.54254940 -1.64450803 L H-HC-0.060300 0 7.17236399 -7.08454940 -1.46550803 L H-HC-0.060300 0 5.62336399 -6.77654940 -0.68050803 L H-HC-0.060300 0 6.74436399 -7.95054940 0.00949197 L N-N- 0 3.48336399 -7.89454940 -1.15250803 L C-CT- 0 2.08936399 -8.10854940 -0.66650803 L C-C- 0 1.35636399 -9.01354940 -1.64550803 L O-O- 0 0.67236399 -9.94054940 -1.26750803 L C-CT- 0 1.35836399 -6.77154940 -0.58450803 L C-CT 0 1.44936399 -6.03754940 -1.92550803 L C-C- 0 0.98036399 -4.59254940 -1.75050803 L O-O2- 0 1.76836399 -3.70154940 -2.02150803 L O-OH- 0 -0.30363601 -4.38854940 -1.29150803 L H-H- 0 3.78536399 -6.99854940 -1.40350803 L
That can correspond to some specific amino acid which are not well defined in the AMBER data base, or it can correspond to some unexpected sequences of atoms (in the case of a chromophore for example because it is not a specific amino acid, GaussView is not able to assign some charges).
If the charges missing correspond to an amino acid not well defined
In this case, have a look at the geometry and at the sequence of atoms, define the amino acid correspond and find in the AMBER paper the charges correspond to this case.
If you can not find a similar sequene in the AMBER paper Most of the time it is the chromophore that is not defined (unexpected sequence of atoms).
I do not have an answer for the moment... It seems that Amber=UnCharged could be a good keyword, but I did not succeed running the calculation...
We'll see......
Files
I put different files here if you want to run some test calculations.
PYP with ONIOM, 2 layers (chromophore + surroundings) : Media:oniom_3phy_pyp_ready.gjf
Optimisation
Now that you have the complete input file, run an optimisation on the structure:
#p opt=quadmac oniom(ub3lyp/6-31g(d):amber=hardfirst)=embedcharge geom=connectivity
Opt=Quadmac is mainly used for problem convergence cases, which does a quadratic step in the coordinates of all the atoms. Such an optimization can use either an updated approximate Hessian for the internal coordinates or an analytically computed Hessian. It computes products of the low-level (MM) Hessian with vectors as needed.
You must specify a ub3lyp method, the default rb3lyp method is not compatible with the system.
This job will terminate without completion:
Error termination request processed by link 9999. Error termination via Lnk1e in /home/gaussian-devel/gaussiandvh01_pgi_725/gdv/l9999.exe at Mon Aug 3 22:16:14 2009.
On inspection of the error message and of the optimisation route in GaussView, it is apparent that the job ran out of steps and was fluctuating between two points. To fix this, copy the last geometry into a new input file and specify a smaller step size:
#p opt=(quadmac,maxstep=10) oniom(ub3lyp/6-31g(d):amber=hardfirst)=embedcharge geom=connectivity
This job completes normally.
Input and output fair
Media:3PHYopt1.gjf
Media:3PHYopt2.gjf
Final geometry: Media:DONE.log
Frequency
Freq, Freq=HPModes
Check that the optimised structure is really a minimum by running a joint optimisation and frequency calculation:
#p opt=(quadmac,maxstep=10) freq oniom(ub3lyp/6-31g(d):amber=hardfirst)=embedcharge geom=connectivity
The geometry modifies slightly: Media:geometry.log
There are no imaginary frequencies, therefore this structure is a minimum.
Low frequencies --- -0.3097 -0.1667 -0.0006 0.0004 0.0006 0.1446
Low frequencies --- 4.5860 5.7688 7.1004
Diagonal vibrational polarizability:
5570.9936676 5790.9109142 7396.2535226
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 4.5858 5.7681 7.1004
Red. masses -- 5.6257 7.1713 6.7230
Frc consts -- 0.0001 0.0001 0.0002
%ModelSys -- 0.8136 5.6385 1.5914
%RealSys -- 99.1864 94.3615 98.4086
IR Inten -- 0.1580 0.7010 0.7201
Atom AN X Y Z X Y Z X Y Z
1 7 0.02 0.01 0.03 0.00 0.00 -0.01 0.02 0.01 0.02
2 6 0.02 0.01 0.03 0.00 0.01 -0.02 0.02 0.00 0.04
3 6 0.02 0.01 0.03 0.00 0.01 -0.01 0.02 0.00 0.04
4 8 0.03 0.01 0.04 0.00 0.00 -0.02 0.02 0.01 0.05
5 6 0.02 0.01 0.05 0.00 0.00 -0.02 0.02 0.01 0.05
As a tip for ease in .log file navigation, use X11 to open the .log files with the "nedit" command. To copy and paste information from a .log to a .com using X11:
1. Open two X11 terminal windows, login as ssh –Y etc… 2. After navigating to the respective directories, open the two files with with cmd: nedit name_1.log & and nedit name_2.com 3. Grab an area using the mouse and copy with ctrl+c (not cmd+c on the mac) from .log, paste with ctrl+v to .com, save
Now copy the same geometry into another input file (to avoid possible corruption with .chk) and run a freq=HPmodes calculation. This keyword gives the vibrational frequency eigenvectors to 5 figures (high precision):
Low frequencies --- -0.3101 -0.1651 -0.0004 0.0001 0.0006 0.1435
Low frequencies --- 4.5908 5.7674 7.1007
Diagonal vibrational polarizability:
5570.6337105 5790.9612763 7395.8674830
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3 4 5
A A A A A
Frequencies --- 4.5906 5.7668 7.1006 7.3648 7.6765
Reduced masses --- 5.6278 7.1696 6.7235 7.0975 7.2004
Force constants --- 0.0001 0.0001 0.0002 0.0002 0.0002
Percent ModelSys --- 0.8203 5.6428 1.5962 0.1342 2.1399
Percent RealSys --- 99.1797 94.3572 98.4038 99.8658 97.8601
IR Intensities --- 0.1579 0.7002 0.7215 0.1148 0.4400
Coord Atom Element:
1 1 7 0.02278 -0.00267 0.02068 0.01019 0.03594
2 1 7 0.01230 0.00471 0.00633 0.05492 -0.00137
3 1 7 0.02519 -0.01023 0.02441 0.07149 -0.00477
Note that both of these .log files are very large (in particular the second one which is 915mb with over 12.7 million lines). Consequently your system may crash if too many processes are running at once.
Freq=ModelModes
Run a freq=modelmodes calculation on the same geometry.
#p freq=(modelmodes) oniom(ub3lyp/6-31g(d):amber=hardfirst)=embedcharge geom=connectivity
This keyword displays modes associated with the smallest model system in this ONIOM calculation:
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1180 1301 1592
A A A
Frequencies -- 347.2729 400.0784 528.7142
Only 31 modes instead of 45? (Would it be possible that Gaussian erase some modes because the file was too big ??)
Freq=(HPModes,ModelModes)
Finally run a freq=(HPmodes,modelmodes) calculation:
#p freq=(hpmodes,modelmodes) oniom(ub3lyp/6-31g(d):amber=hardfirst)=embedcharge geom=connectivity
In comparison, the last two files are much smaller than the first two and therefore easier to work with.
If you compute the calculation with Freq=(HPModes,ModelModes) keyword you get the information that you need directly:
- First, you know if you have some imaginary frequencies (there is no mention of imaginary frequencies in the .log if there aren't any).
Full mass-weighted force constant matrix:
Low frequencies --- -727.1752 -603.3920 -441.6366 -348.9122 -336.0877 -330.3771
Low frequencies --- -318.1434 -313.7065 -289.1518
****** 362 imaginary frequencies (negative Signs) ******
Diagonal vibrational polarizability:
52770.5393600 27276.1819608 39738.0499572
NorSel: MapVib= 2181 2332 2360 2364 2521 2847 3397 3653 3874 4190
NorSel: MapVib= 4714 4764 4822 4839 5543 5580 5585 5736 5765 5766
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
2181 2332 2360
A A A
Frequencies -- 781.7748 858.7045 871.9542
Red. masses -- 4.5651 1.7393 1.9629
- Second, the precision will be better, because the HPModes keyword enable you to get 5 figures for the vibrational frequency eigenvectors, which could be useful so as to run some other calculation to et some overlap.
Input fair
Media:3PHYopt1_1freq.gjf
Media:3PHYopt1_1freq2.gjf
Media:3PHYopt1_1freq3.gjf
Media:3PHYopt1_1freq4.gjf
Media:3PHYopt1_1freqMM.gjf
Back to ONIOM for biomolecules
Back to ONIOM tutorial (G03)