Od416
NH3 Optimisation
What is the molecule? NH3
What is the calculation method? RB3LYP
What is the basis set? 6-31G(d,p)
What is the final energy E(RB3LYP) in atomic units (au)? -56.55776873
What is the RMS gradient? 0.00000485
What is the point group of your molecule? C3V
Bond Length: 1.01798 au
Bond Angle: 105.741 degrees
Item Table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000070 0.001800 YES
RMS Displacement 0.000033 0.001200 YES
Predicted change in Energy=-5.785197D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log File
Static Image
Jmol Image
NH3 |
Display Vibrations File
Display Vibrations Questions
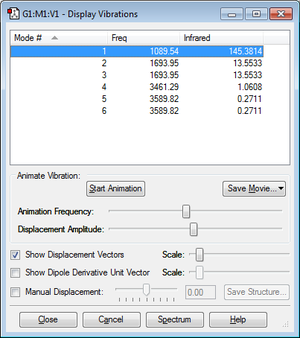
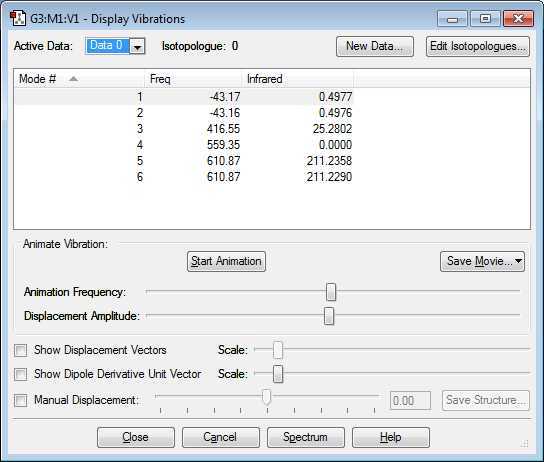
how many modes do you expect from the 3N-6 rule? 3 modes because there are 3 atoms and 3(3) - 6 = 3
which modes are degenerate (ie have the same energy)? modes 2 and 3; modes 5 and 6
which modes are "bending" vibrations and which are "bond stretch" vibrations?
mode 1 is bending
mode 2 is bending
mode 3 is bending
mode 4 is stretching
mode 5 is stretching
mode 6 is stretching
which mode is highly symmetric?
mode 4 is highly symmetric
one mode is known as the "umbrella" mode, which one is this?
mode 1 is the umbrella mode
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
there would be 3 bands on an an experimental spectrum of gaseous ammonia
Charge Distributions
The N atom has a -1.125 charge and the H atom has a +0.375 charge
N2 Optimisation
What is the molecule? N2
What is the calculation method? RB3LYP
What is the basis set? 6-31G(d,p)
What is the final energy E(RB3LYP) in atomic units (au)? -109.52412868
What is the RMS gradient? 0.00000001
What is the point group of your molecule? D∞h
Bond Length: 1.10550 au
Bond Angle: 180 degrees
Item Table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-9.123258D-16
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log File
Static Image
Jmol Image
N2 |
Display Vibrations File
none are negative
H2 Optimisation
What is the molecule? H2
What is the calculation method? RB3LYP
What is the basis set? 6-31G(d,p)
What is the final energy E(RB3LYP) in atomic units (au)? -1.17853936
What is the RMS gradient? 0.00000001
What is the point group of your molecule? D∞h
Bond Length: 0.74279 au Bond Angle: 180 degrees
Item Table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.167770D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log File
Static Image
Jmol Image
H2 |
Display Vibrations File
Reactivity Questions
E(NH3)= -56.55776873 au
2*E(NH3)= -113.11553746 au
E(N2)= -109.52412868 au
E(H2)= -1.17853936 au
3*E(H2)= -3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=
-113.11553746 - (-109.52412868 -3.53561808) = -0.05579070 au = -146.47848285 kj/mol
the product (NH3) is more stable than the reactants (H2 and N2)
F2 Optimisation
What is the molecule? F2
What is the calculation method? RB3LYP
What is the basis set? 6-31G(d,p)
What is the final energy E(RB3LYP) in atomic units (au)? -199.49825218
What is the RMS gradient? 0.00007365
What is the point group of your molecule? D∞h
Bond Length: 1.4028 au
Bond Angle: 180 degrees
Item Table
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000157 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-2.131104D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4028 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log File
Static Image
Jmol Image
F2 |
Display Vibrations File
Charge Distribution Info:
Both F atoms have a 0.000 charge (molecule is non-polar)
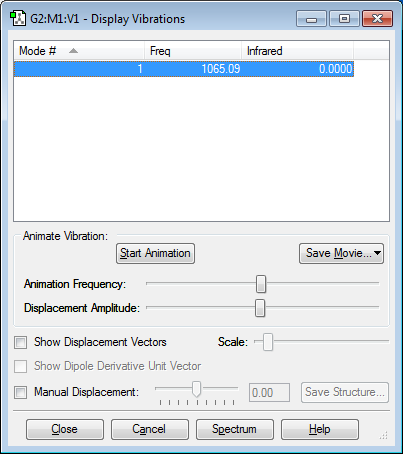
Vibration Info:
This molecule has 1 vibrational mode as predicted by the 3N - 5 rule for linear molecules (3*2-5 = 1). It is a symmetric stretch with the frequency 1065.09
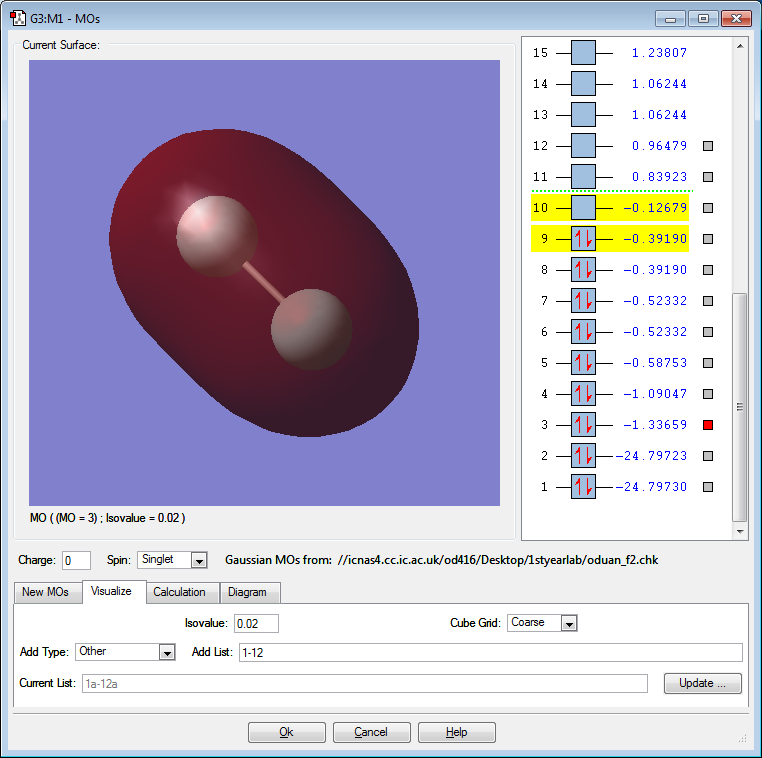
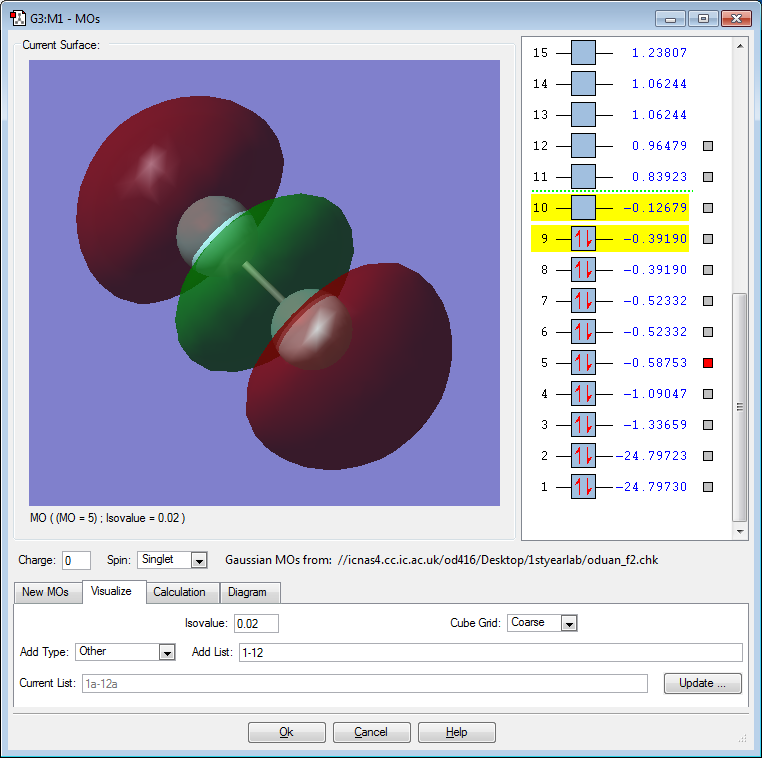
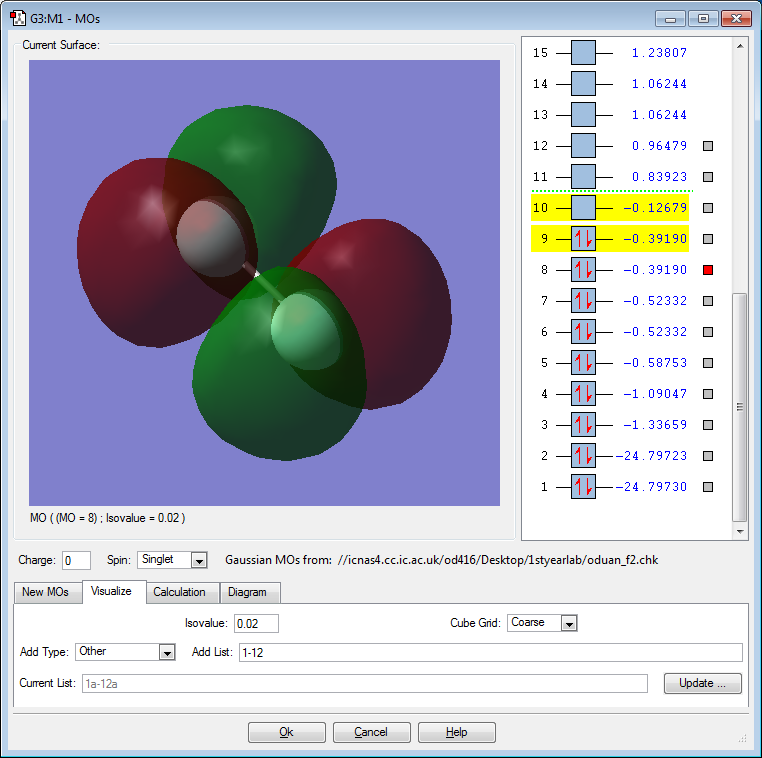
F2 Orbital Pictures and Discussion
The above orbital is a bonding 1σ orbital created by the constructive overlap of the 1s atomic orbital on each F atom. The MO is deep in energy. The MO is occupied. This MO is not directly involved in bonding.
The above orbital is an antibonding 1σ* orbital created by the destructive overlap of the 1s atomic orbital on each F atom. The MO is deep in energy. The MO is occupied. This MO is not directly involved in bonding. This MO effectively "cancels out" the 1σ orbital.
The above orbital is a bonding 2σ orbital created by the constructive, head-on overlap of the 2p atomic orbital on each F atom. The MO is deep in energy. The MO is occupied. This MO is responsible for the single bond between the F atoms.
The above orbital is an bonding 2π orbital created by the sideways, constructive overlap of the 2p atomic orbital on each F atom. The MO is deep in energy. The MO is occupied. This bonding orbital contributes positively to the overall stability of the atom.
The above orbital is an antibonding 2π* orbital created by the sideways, destructive overlap of the 2p orbital on each F atom. The MO is the HOMO. The MO is occupied. Electrons from this molecule will interact with the LUMO of another atom in reactions to potentially form new bonds.
Independent Work
For my independent work I decided to run an analysis on ClF3.
What is the molecule? ClF3
What is the calculation method? RB3LYP
What is the basis set? 6-31G(d,p)
What is the final energy E(RB3LYP) in atomic units (au)? -759.44149573
What is the RMS gradient? 0.00000553
What is the point group of your molecule? C3V
Bond Length: 1.75684 au
Bond Angle: 120 degrees
Item Table
Item Value Threshold Converged?
Maximum Force 0.000011 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.000049 0.001800 YES
RMS Displacement 0.000032 0.001200 YES
Predicted change in Energy=-8.249258D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.7568 -DE/DX = 0.0 !
! R2 R(1,3) 1.7568 -DE/DX = 0.0 !
! R3 R(1,4) 1.7568 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log File
Static Image
Jmol Image
ClF3 |
Display Vibrations File
Vibrational Information
Mode 1 is bending (scissor)
Mode 2 is bending (rocking)
Mode 3 is bending (umbrella)
Mode 4 is symmetric stretching
Modes 5 and 6 are asymmetric stretching
Modes 5 and 6 are degenerate
Charge Information
The Cl atom has a + 0.898 charge
The F atoms have a -0.338 charge