ModelLab2
Optimised NH3 molecule
Gaussian Calculation Summary
Calculation method : RB3LYP
Basis set : 6-31G(d.p)
Final energy E(RB3LYP) : -56.55776873 a.m.u.
RMS Gradient Norm : 0.00000485 a.m.u.
The point group of NH3 : C3V
Bond length of N-H bond = 1.01798 Angstrom
Bond angle of H-N-H = 105.741 Degrees
Image of the molecule
NH3 molecule |
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:NH3Optimisationkbg15.LOG
"Item" table from log file
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986267D-10
Optimization completed.
-- Stationary point found.
Display Vibrations window
IR analysis
According to the 3N-6 rules, N is the number of atoms and it's 4 in this case, I should have 6 distinct vibrational modes. Mode 2 and Mode 3 are degenerate which means they have the same energy. Mode 4,5 and 6 are degenerate too. Mode 4, 5 and 6 are stretches mode 1,2 and 3 are bending vibrations. Mode 4 is highly symmetric. Mode 1 is the "umbrella" mode. I will get 2 bands in the experimental spectrum which indicates mode 1 , mode 2 and mode 3. Mode 2 and 3 has similar frequency hence at the will be seen as 1 band. IR intensities of band 4,5 and 6 are too low to be detected.

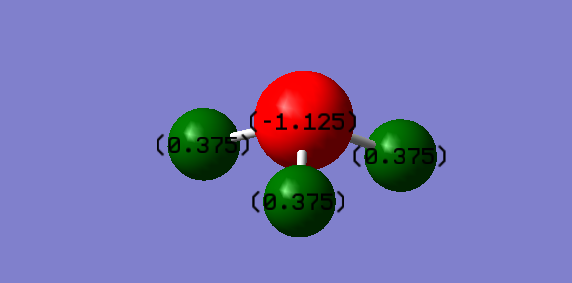
Atomic charges
Charge on N = -1.125 Charge on both H's = 0.375 Nitrogen atom is more electronegative than Hydrogen atom. Electrons are attracted towards N causing it to have a negative charge and hence hydrogen is electron deficient and hence positively charged.
Optimised H2 molecule
Gaussian Calculation Summary
Calculation method : RB3LYP
Basis set : 6-31G(d.p)
Final energy E(RB3LYP) :-1.17853936 a.m.u.
RMS Gradient Norm : 0.00000017 a.m.u.
The point group of H2 : D ∞h
Image of the molecule
H2 molecule |
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:H2_OPTIMISATION_kbg15.LOG
"Item" table from log file
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13 Optimization completed. -- Stationary point found.
Vibration Frequency
The Vibration Frequency of H2 molecule is 4465.68 cm-1. The molecule is not IR active because there is not change of dipole when the molecule vibrates.
Optimised N2 molecule
Gaussian Calculation Summary
Calculation method : RB3LYP
Basis set : 6-31G(d.p)
Final energy E(RB3LYP) : -109.52412868 a.m.u.
The point group of N2 : D ∞h
Image of the molecule
N2 molecule |
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:N2_OPTIMISATIONkbg15.LOG
"Item" table from log file
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401103D-13
Optimization completed.
-- Stationary point found.
Vibration Frequency
The Vibration Frequency of N2 molecule is 2457.33 cm-1. The molecule is not IR active because it doesn't change its dipole upon vibration.
Reaction Between N2 and H2 forming NH3
E(NH3)= -56.55776873 a.m.u.
2*E(NH3)= -113.115537 a.m.u.
E(N2)= -109.52412868 a.m.u.
E(H2)= -1.17853936 a.m.u.
3*E(H2)=-3.53561808 a.m.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579024 a.m.u. = -146.48 kJ/mol
Gaseous product is more stable hence the reaction is exothermic.
Molecule of my choice (CO)
Gaussian Calculation Summary
Calculation method : RB3LYP
Basis set : 6-31G(d.p)
Final energy E(RB3LYP) : -113.30945314 a.m.u.
RMS Gradient Norm : 0.00001828 a.m.u.
The point group of CO : D∞h
Bond length of C≡O = 1.13793 Angstrom
Image of the molecule
CO molecule |
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:COkbg15.LOG
"Item" table from log file
Item Value Threshold Converged?
Maximum Force 0.000032 0.000450 YES
RMS Force 0.000032 0.000300 YES
Maximum Displacement 0.000012 0.001800 YES
RMS Displacement 0.000018 0.001200 YES
Predicted change in Energy=-3.956716D-10
Optimization completed.
-- Stationary point found.
Frequency Analysis
Linear molecule has 3N-5 numbers of vibrational modes. N=2, hence the molecule has 1 vibrational mode. It absorbs the wavenumber 2209.14 cm-1 and this can be observed in IR spectrum.

Atomic charges
Charge on C = 0.506 Charge on O = -0.506 Oxygen atom is more electronegative than carbon atom. Electrons are attracted towards O causing it to have a negative charge and hence C is electron deficient and hence positively charged.

Molecular Orbitals of CO

The Lowest Unoccupied Molecular Orbital (LUMO) is the π* orbital of CO.

The Highest Occupied Molecular Orbital (HOMO) of CO is the sigma bonding orbital. π* has relative energy of -0.02177 but σ3 has relative energy of -0.37145. Hence, there is a huge gap between the LUMO and HOMO. The huge electron cloud around C signifies the nucleophilic site is at C, not O.

The following filled orbitals are the pi bonding orbitals. They have relative energies of -0.46743 which are not too much difference compared to -0.2177. Smaller gap between σ3 and π.

The anti-sigma orbital has the relative energy of -0.57004 and is quite close in energy to the π orbitals, hence a small gap is observed.

The σ1 orbital has relative energy of -1.15791 has a significant drop in energy from the σ2, large gap is observed. Note that the gap is not as big as the one between LUMO and HOMO.

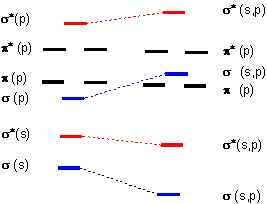
Mixing of the σs with σp and σ*s with σ*p is explained using the diagram below. This explains why σ3 is higher in energy than the two π orbitals and the energy gaps between all the MO's. Basically, molecular orbitals with the same geometry can mix together. For example, 2σg and 3σg can mix together resulting a lower energy new σ2 orbital and higher energy σ3. Same thing happens to the antibonding sigma orbitals.
Independence
Ethanal molecule
Calculation method : RB3LYP
Basis set : 6-31G(d.p)
Final energy E(RB3LYP) : -153.83392903 a.m.u.
RMS Gradient Norm : 0.00018263 a.m.u.
Dipole moment : 2.5327 Debye
The point group of NH3 : C1
Image of the molecule
NH3 molecule |
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:ETHANALkbg15.LOG
"Item" table from log file
Item Value Threshold Converged?
Maximum Force 0.000393 0.000450 YES
RMS Force 0.000131 0.000300 YES
Maximum Displacement 0.001239 0.001800 YES
RMS Displacement 0.000721 0.001200 YES
Predicted change in Energy=-9.663621D-07
Optimization completed.
-- Stationary point found.
IR analysis
According to the 3N-6 rules, N is the number of atoms and it's 7 in this case, I should have 15 distinct vibrational modes. The molecule is IR active. IR spectrum is predicted as diagram below.

Atomic charges
Charge on O = -0.523 Charge on Carbonyl C = 0.412 Charge on Hydrogen attached to carbonyl carbon = 0.142 Charge on alpha carbon = -0.802 Charge on Hydrogen attached to the alpha carbon = 0.265/0.241 This shows that the alpha carbon is negatively charged and hence can be a potential nucleophilic site besides the oxygen atom. Alpha carbon is even more negatively charged than the oxygen. Note that the hydrogen is quite positively charged and hence the molecule could be deprotonated using a strong base. Molecular orbitals need to be considered to know the site of reaction.

Molecular orbitals of Ethanal
Diagram below shows the LUMO of ethanal. This shows where the electrons will be first filled in by a nucleophile. The huge electron cloud at the carbonyl carbon indicates the electrophilic site of the molecule.

As we know, ethanal is a electrophile when it is in its protonated form. But when it is deprotonated, it is a nucleophile, (enolate ion) and both Oxygen and Alpha carbon is the nucleophilic site.

