Modcej17
Computational Lab 2
Molecule: NH3
Bond Length: 1.01798 a.u.
Bond Angle: 105.745
Calculation Method: B3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -56.55776873 au
RMS Gradient: 0.00000323 au
Point Group: C3V
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000014 0.001800 YES RMS Displacement 0.000009 0.001200 YES Predicted change in Energy=-1.166745D-10
NH |
NH3 Vibrational Modes

| Frequency | IR intensity | Vibrational mode |
|---|---|---|
| 1089 | 145 | A1 (1) |
| 1693 | 13 | E (2) |
| 1693 | 13 | E (3) |
| 3461 | 1 | A1 (4) |
| 3589 | 0 | E (5) |
| 3589 | 0 | E (6) |
NH3 Charge Distribution
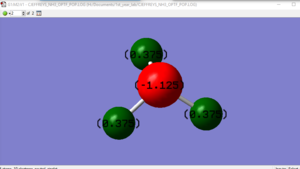
Questions
how many modes do you expect from the 3N-6 rule?
3*4-6=6
which modes are degenerate (ie have the same energy)?
E(2) and E(3) are degenerate and E(3) and E(4) are degenerate
which modes are "bending" vibrations and which are "bond stretch" vibrations?
A1(1), E(2) and E(3) are all bending, A1(4), E(5) and E(6) are all bond stretch
which mode is highly symmetric?
A1(1) one mode is known as the "umbrella" mode, which one is this?
A1(1)
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2
Molecule: N2
Bond Length:1.106
Calculation Method: RB3LYP
Basis Set: 6-31G(D,P)
Final Energy E(RB3LYP): -109.524 a.u.
RMS Gradient: 0.00000060 a.u.
Point Group: D*H
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401020D-13
N |
N2 Vibrational Modes
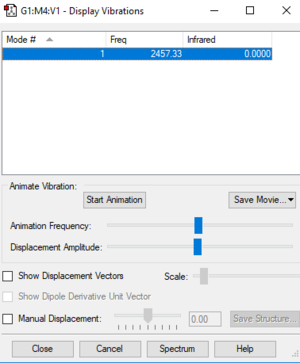
| Frequency | IR intensity | Vibrational mode |
|---|---|---|
| 2457 | 0 | SGG |
N2 Charge Distribution
Questions
Nitrogen has 1 mode of vibration (bond stretch)
how many bands would you expect to see in an experimental spectrum of gaseous Nitrogen?
none as it is IR inactive
Unique identifier code: DEKFUX
https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=DEKFUX&DatabaseToSearch=Published
N-N bond length in structure: 1.086 a.u.
Bond lengths are determined by the amount of electron density between the two nuclei of the atoms involved. Increased electron density causes a shorter bond as the positive nuclei are more strongly attracted to the increased negative charged of the electron cloud. in this crystal structure, we see the dinitrogen species bound to a Ru metal ion, which acts as an electron withdrawing species. Due to electrons being withdrawn from the dinitrogen, there is less negative character between the nuclei and thus the bond length increases. This is consistent with the calculated result on Gaussian (1.106 a.u.) compared to the observed result from the crystal structure (1.086 a.u.).
Molecule: H2
Bond Length: 0.6 au
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -1.17853936 au
RMS Gradient: 0.00000017 au
Point Group: D*H
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13
NH |
H2 Vibrational Modes
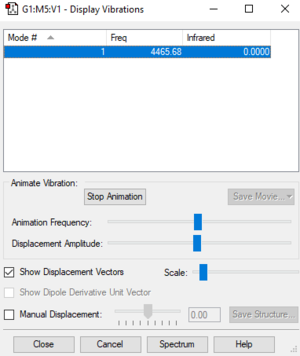
| Frequency | IR intensity | Vibrational mode |
|---|---|---|
| 4466 | 0 | SGG |
H2 Charge Distribution
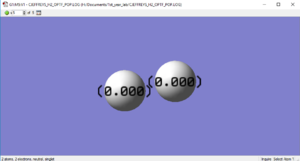
Questions
Hydrogen has 1 mode of vibration (bond stretch)
how many bands would you expect to see in an experimental spectrum of gaseous Hydrogen?
none as it is IR inactive
Haber Process
NH3 enthalpy change: -148492 kJmol-1
2*NH3 enthalpy change: -296984.865017 kJmol-1
N2 enthalpy change: -287555.284 kJmol-1
H2 enthalpy change: -3094.25532539 kJmol-1
3*H2 enthalpy change: -9282.765269 kJmol-1
ΔHtotal = 2*NH3 - (N2 + 3*H2)
=-146.81 kJmol-1
The ammonia product is more stable because it has a negative enthalpy change which is favourable in Gibbs Free energy.
Molecule of choice: NF3
Bond Length: 1.38404 a.u.
Bond Angle: 101.830
Calculation Method: B3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -354.07131058 a.u.
RMS Gradient: 0.00010256 a.u.
Point Group: C3V
Item Value Threshold Converged? Maximum Force 0.000164 0.000450 YES RMS Force 0.000108 0.000300 YES Maximum Displacement 0.000612 0.001800 YES RMS Displacement 0.000296 0.001200 YES Predicted change in Energy=-1.274067D-07
NH |
NF3 Vibrational Modes
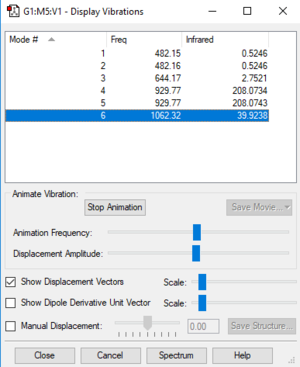
| Frequency | IR intensity | Vibrational mode |
|---|---|---|
| 482 | 1 | E (1) |
| 482 | 1 | E (2) |
| 644 | 3 | A1 (3) |
| 929 | 208 | E (4) |
| 929 | 208 | E (5) |
| 1062 | 40 | A1 (6) |
NF3Charge Distribution
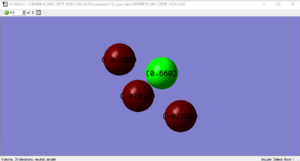
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES - however, all your jmols are labelled 'NH' which is not informative at all.
NH3 0/1
Have you completed the calculation and given a link to the file?
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
NO
Have you included answers to the questions about vibrations and charges in the lab script?
YES - you answered all questions the right way. Well done! You could have explained the charges based on an electronegativity argument.
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1. Additionally, you could have explained that the charges are 0 as the electronegativities are equal.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 2/5
Have you completed the calculation and included all relevant information?
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Have you added information about MOs and charges on atoms?
You displayed the charges and vibrations correctly but missed to explain your results. You missed to describe which orbitals are occupied/unoccupied, bonding/anti-bonding/non-bonding and to comment on their relative energies. In most cases you correctly described the contributions of the Mos to the MOs. Instead of only referring to s and p orbitals you should have included the principle quantum number of each orbital to make your discussion easier to follow. You stated the energies of the orbitals but messed up the numbers for the third and fourth displayed MO. You missed to to say which MO is bonding/anti-bonding/non-bonding and occupied/unoccupied. You stated p AOs to be anti-bonding. An AO itself has no bonding/anti-bonding character.
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far
NO - No independent work has been identified.





