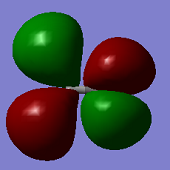
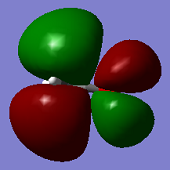
Mod im915 01096773
Molecular Modelling
NH3 Molecule
| N-H bond length | H-N-H bond angle | Calculation Method | Basis set | Final energy E(RB3LYP)(a.u.) | Point Group |
|---|---|---|---|---|---|
| 1.01798 | 105.741 | B3LYP | 6-31G(d,p) | -56.55776873 | C3v |
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
NH3 molecule |
Log file for NH3 here
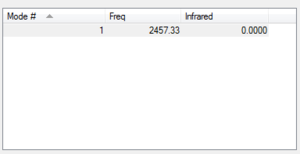
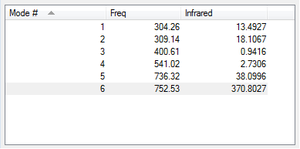
Vibrations
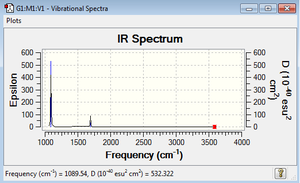
From the 3N-6 rule, 6 vibrations would be expected as seen (N = 4, 3N-6 = 12-6) There are 2 pairs of degenerate modes- modes 2 and 3, and modes 5 and 6 as named in the above image
Modes 1,2 and 3 are 'bending' vibrations, modes 4,5 and 6 are 'bond stretching' vibrations
Mode 4 is the highly symmetric mode as it retains all symmetry elements
Mode 1 is the umbrella mode
In an experimental spectrum, you would expect to see 2 bands in the spectrum of gaseous ammonia- while there are 4 adsorptions, modes 4, 5 and 6 give very small absorbance that on a standard spectrometer would not be detected.
Charges
The charge of the N atom is -1.125 and the charge on each H atom is +0.375. This gives a net charge of 0 over the whole atom. You would expect the N atom to be negative as it is more electronegative than the H atoms, so it will pull the electron density towards itself.
The Haber-Bosch process
In the Haber-Bosch process, nitrogen (from the air) and hydrogen (derived mainly from natural gas i.e. methane) are combined to give ammonia. The reaction is reversible.
N2 + 3H2 ⇌ 2NH3
N2 Molecule
| N-N bond length | Calculation Method | Basis set | Final energy E(RB3LYP)(a.u.) | Point Group |
|---|---|---|---|---|
| 1.10550 | B3LYP | 6-31G(d,p) | -109.52412868 | D∞h |
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
N2 molecule |
Log file for N2 here
H2 Molecule
| H-H bond length | Calculation Method | Basis set | Final energy E(RB3LYP)(a.u.) | Point Group |
|---|---|---|---|---|
| 0.74279 | B3LYP | 6-31G(d,p) | -1.17853936 | D∞h |
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES

H2 molecule |
Log file for H2 here
Calculations
E(NH3)= -56.55776873 au
2*E(NH3)= -113.11553746 au
E(N2)= -109.52412868 au
E(H2)= -1.17853936 au
3*E(H2)= -3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557907 au
Total energy for reaction converting N2 + 3H2 ⇌ 2NH3 => -146.47849401kJ/mol-1
So the ammonia product is more stable as the energy change calculated is negative, so the reaction is exothermic.
Small molecule of own choice- ClF3 Molecule
| Cl-F bond length (axial) | Cl-F bond length (equatorial) | F-Cl-F bond angle | Calculation Method | Basis set | Final energy E(RB3LYP) (a.u.) | Point Group |
|---|---|---|---|---|---|---|
| 1.72852 | 1.65118 | 87.193 | B3LYP | 6-31G(d,p) | -759.46531666 | C2v |
Item Value Threshold Converged? Maximum Force 0.000193 0.000450 YES RMS Force 0.000096 0.000300 YES Maximum Displacement 0.001280 0.001800 YES RMS Displacement 0.000808 0.001200 YES
ClF3 molecule |
Log file for ClF3 here
Charges

The charge of the Cl atom is +1.225, the charges on the axial F atoms are -0.454, and the charge of the equatorial F atom is -0.316. You would expect the Cl atom to be positive as it is more electropositive than the F atoms, so the F atoms will pull more electron density towards themselves. The equatorial F atom is less negatively charged than the axial F's, possibly due to less electron density in this Cl-F bond.
Molecular Orbitals
ClF3 has 44 electrons, so will have 22 electron pairs each occupying a different molecular orbital. Shown above are some of the interesting MO's calculated by Gaussian.
The first MO shown (top -left) is a non-bonding orbital, with electron density very deep in to the nucleus of the Cl atom and no electron density located over any of the Cl-F bonds- this is likely from the Cl 1s orbital.
The second MO shown (top-centre) shows full bonding character as electron density (in-phase) is located over the entire molecule. There are no nodes, so this likely comes from in-phase overlap of s orbitals.
The third MO shown (top-right) shows in-phase overlap over the axial F-Cl-F bond, but this electron density is out of phase with the electron density on the third equatorial F atom, and there is no electron density located over this Cl-F bond.
The fourth MO shown (bottom-left) is fully anti-bonding- there is no electron density located between any of the atoms i.e. there is no electron density located over any of the bonds.
The fifth MO shown (bottom-centre) shows electron density above and below the plane of the molecule with a nodal plane in the plane of the molecule. This is likely from in phase overlap of perpendicular p-orbitals from all atoms.
The sixth MO shown (bottom-right) is the highest occupied MO- the HOMO. This shows anti-bonding character- there are no overlaps between in phase electron density between atoms i.e. located over bonds.
Independent extension- Methanal Molecule (Formaldehyde)
| C=O bond length | C-H bond length | Calculation Method | Basis set | Final energy E(RB3LYP)(a.u.) | Point Group |
|---|---|---|---|---|---|
| 1.20663 | 1.11045 | B3LYP | 6-31G(d,p) | -114.50319936 | C2v |
Item Value Threshold Converged? Maximum Force 0.000045 0.000450 YES RMS Force 0.000017 0.000300 YES Maximum Displacement 0.000052 0.001800 YES RMS Displacement 0.000027 0.001200 YES

Methanal molecule |
Log file for Methanal here