ModFEG1908
EX3 Section
BH3 Molecule
B3LYP/6-31G level
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000023 0.001800 YES RMS Displacement 0.000015 0.001200 YES
Frequency Analysis Log File:File:FG BH3 FREQ.LOG
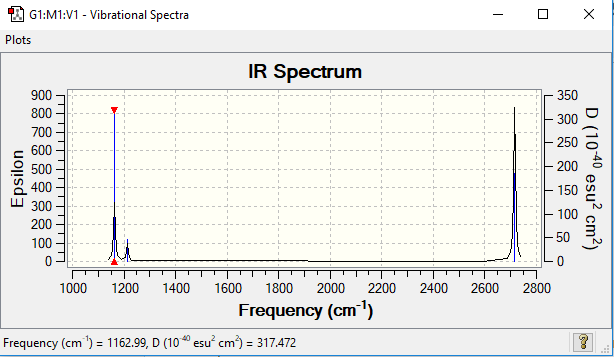
Low frequencies --- -2.3742 -1.0821 -0.0054 2.0837 10.1950 10.2522 Low frequencies --- 1162.9856 1213.1754 1213.1781
Optimised BH3 |
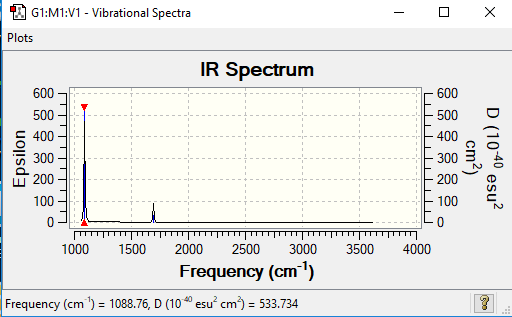
There are 6 vibrations of the BH3 molecule but less than 6 peaks. There is only 3 peaks as there are two pairs of degenerate vibrations and one vibration that is not IR active.
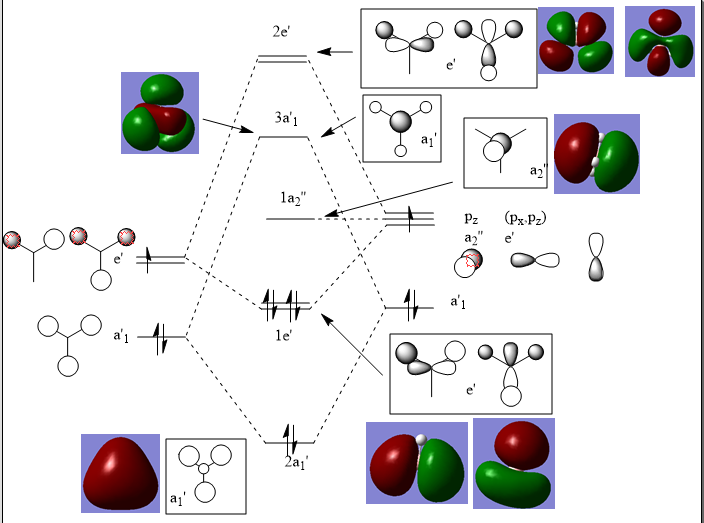
BH3 molecular Orbitals
There is no significant differences between the real and LCAO MOs. This shows that qualitative MO theory is very accurate and useful in this instance. The 1s MO was also created on Gussian but was much deeper in energy and so is not present in the MO diagram.
NH3 Molecule
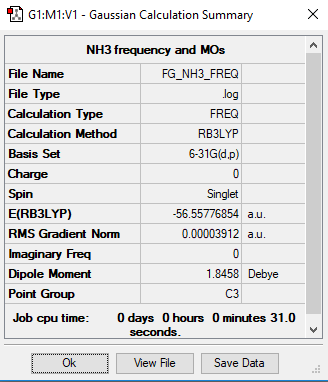
B3LYP/6-31G level
Item Value Threshold Converged?
Maximum Force 0.000060 0.000450 YES
RMS Force 0.000040 0.000300 YES
Maximum Displacement 0.000369 0.001800 YES
RMS Displacement 0.000162 0.001200 YES
Frequency Analysis Log File:File:FG NH3 FREQ.LOG
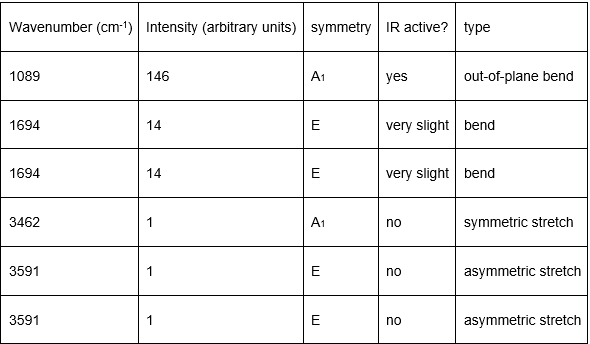
Low frequencies --- -32.4235 -32.4224 -11.4276 -0.0047 0.0113 0.0475 Low frequencies --- 1088.7628 1694.0251 1694.0251
Optimised NH3 |
NH3BH3 Molecule
B3LYP/6-31G level
Ng611 (talk) 20:08, 20 May 2019 (BST) Your energy here is slightly off unfortunately.
Item Value Threshold Converged? Maximum Force 0.000164 0.000450 YES RMS Force 0.000043 0.000300 YES Maximum Displacement 0.001168 0.001800 YES RMS Displacement 0.000259 0.001200 YES
Frequency Analysis Log File:File:FG NH3BH3 FREQ.LOG
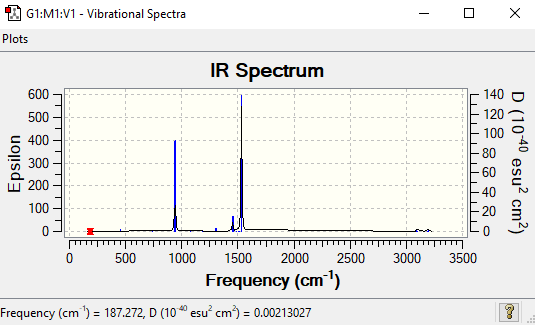
Low frequencies --- -0.0007 -0.0006 -0.0005 0.8656 13.9276 26.6214 Low frequencies --- 266.1273 633.9919 641.7220
Optimised NH3BH3 |
Association Energy
E(NH3)=-56.55776871 a.u. E(BH3)=-26.61532364 a.u. E(NH3BH3)=-83.21320666 a.u.
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)] ΔE=-0.04011431 a.u.=-105 kJ(3.s.f)
Ng611 (talk) 20:10, 20 May 2019 (BST) SLightly off the true value due to incorrect energy for nh3bh3
The B-N dative bond is of medium strength. The value is around 25 kcal/mol dome bonds association values can be as low as 0.002 kcal/mol or as high as 152 kcal/mol.
Ng611 (talk) 20:10, 20 May 2019 (BST) A comparison to known experimental bond enthalpies (preferably isoelectronic) would be ideal here.
NI3
B3LYP/6-31G level
Frequency Analysis Log File: File:FG NI3 FREQ CORRECT.log
Item Value Threshold Converged? Maximum Force 0.000067 0.000450 YES RMS Force 0.000044 0.000300 YES Maximum Displacement 0.000482 0.001800 YES RMS Displacement 0.000327 0.001200 YES
Optimised NI3 |
The optimized distance is 2.184 Angstroms (3.d.p)
Project Section
Ionic Liquids
[N(CH3)4]+ Molecule
B3LYP/6-31G level
Frequency Analysis Log File:File:FG N(CH3)4 FREQ.LOG
Item Value Threshold Converged? Maximum Force 0.000136 0.000450 YES RMS Force 0.000050 0.000300 YES Maximum Displacement 0.000502 0.001800 YES RMS Displacement 0.000173 0.001200 YES
Optimised N(CH3)4 |
Low frequencies --- -15.2627 -11.7593 -0.0010 -0.0008 -0.0006 7.9555 Low frequencies --- 187.2822 288.3342 289.8038
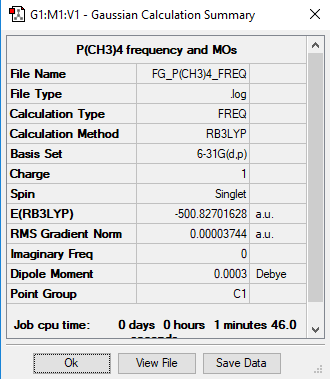
[P(CH3)4]+ Molecule
B3LYP/6-31G level
Frequency Analysis Log File:File:FG P(CH3)4 FREQ.LOG
Item Value Threshold Converged? Maximum Force 0.000067 0.000450 YES RMS Force 0.000016 0.000300 YES Maximum Displacement 0.001236 0.001800 YES RMS Displacement 0.000308 0.001200 YES
Optimised P(CH3)4 |
Low frequencies --- -16.4962 -8.7101 -0.0038 -0.0031 -0.0026 17.0348 Low frequencies --- 152.5604 185.4897 189.0154
Charge Distributions
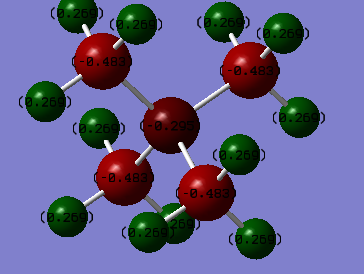
In N(CH3)4+ the charge on the N atom is -0.295 e, the charge on the C atoms is -0.483 e, and the charge on the H atoms is 0.269 e.
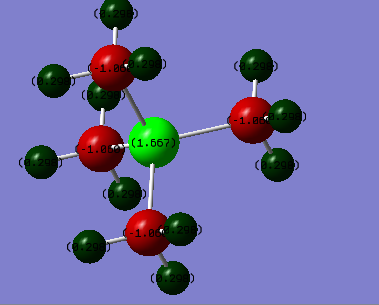
In In P(CH3)4+ the charge on the P atom is 1.667 e, the charge on the C atoms is -1.060 e, and the charge on the H atoms is 0.298 e.
The most notable difference between the two molecules' charge distribution is that the phosphorous atom has a positive charge and the Nitrogen atom has a negative charge. The magnitude of the charge on phosphorous was also larger at 1.667 e compared to the -0.483 e on the nitrogen. The carbons on both molecules were more negative than the central atoms, with a -1.060 e charge for the phosphorous and a -0.483 e for the Nitrogen complex. The hydrogens both had similar positive charges on the hydrogens with a slightly more positive vealue on the phosphorous complex at 0.298 e compared to the 0.269 e on the nitrogen one. The difference in charges in the complexes is probably due to the fact that the nitrogen atom is more electronegative than the phosphorous atom. THis means that electrons are pulled away from the carbons in the nitrogen complex more strongly making the carbon atoms less negatively charged and the nitrogen more negatively charged.
Ng611 (talk) 20:17, 20 May 2019 (BST) You should also consider the effect of molecular symmetry.
In the [NR4]+ depiction the formal positive charge is placed on the central nitrogen atom implying that the charge distribution on this atom should be positive. From the calculations for the Nitrogen complex the charge on the nitrogen atom was -0.295 e and the positive charge was on the hydrogen atoms with a value of 0.269 e.
Ng611 (talk) 20:17, 20 May 2019 (BST) Yes but why is this the case?
Molecular Orbitals

Ng611 (talk) 20:22, 20 May 2019 (BST) Good LCAO analysis. You should consider labelling the key interactions for your MOs though.
In MO 10 the s orbital on the nitrogen atom is anti-bonding with the methyl ligand molecular orbitals, which are all bonding with each other so have been represented as s orbitals.
In MO 15 the central nitrogen atom is non-bonding with the methyl groups. The methyl group ligand molecular orbitals have been represented as p orbitals for simplification. These p orbitals bond and antibond with each other as they overlap.