Mod:gv advanced
Advanced GaussView Building Tutorial
There are several useful options available in GaussView to help build and manipulate large molecules. In these tutorials, we cover some of the less standard options available in the package.
Appending a structure
Combining two structures together that have been built and/or optimized separately can be useful for building dimers or docking substrates into proteins.
Click here for the tutorial on Building Tyrian Purple Dye as an example of how to append a structure.
Creating a custom fragment
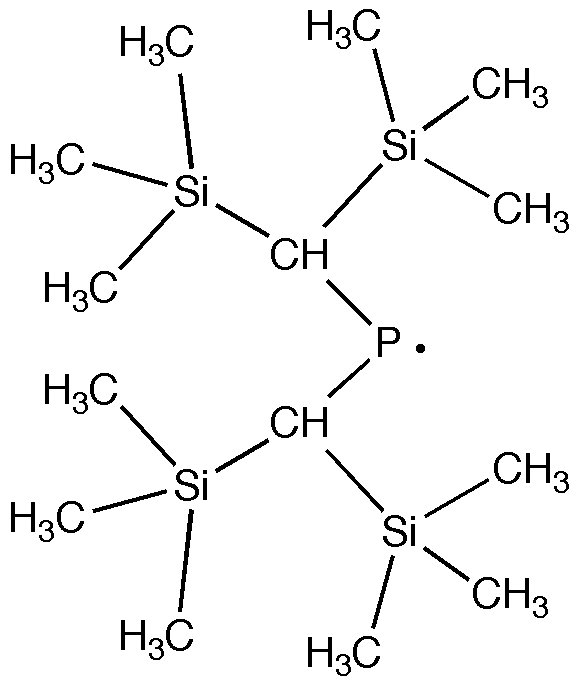
You can build your own fragment in GaussView and save it to the Custom Fragment Library which can save time if your molecule has several bulky ligands that have the same structure.
(1) Build the Si(CH3)3 fragment.
(2) Add the created fragment to the Custom Fragment Library by following the instructions given here: Custom Fragment Library.
(3) Build the following radical using the custom fragment created in (2) and save the structure to a file.
(4) Optimize the radical using PM3. You are advised NOT to use the Clean function first in this case as it tends to result in overlapping methyl groups.
(5) The above radical dimerises. Open the same radical again in the same window by selecting Append all files to active molecule when you open the output file generated in (4). Now build the dimer. See if you can optimize it at the PM3 level also.
Editing files in GaussView
There are limitations in GaussView for setting up certain Gaussian jobs. An example of this is when one would like to change the isotope of an atom in a frequency calculation. Here we look at the effect of substituting D for H in methane.
(1) Draw a methane molecule and set up an optimization and frequency calculation at the HF/3-21G level. Run the calculation and visualize the vibrational frequencies.
(2) Starting from the optimized structure, set up a new frequency calculation at the same level of theory. Add the keyword Freq=ReadIsotopes in the Additional Keywords field. Click Update.
(3) We will set up a frequency calculation at a different temperature (200K) and at 1 atmospheric pressure, replacing one of the hydrogen atoms with deuterium. Click on the Edit button. You will be prompted to save the file. The input file will then be opened in WordPad.
(4) Now edit the input file manually to add the temperature, atmospheric pressure and isotopes as shown below (comments are in red). These should be listed at the end of the input file after the molecule specification separated by a blank line. Remember to leave a blank line at the end of the input file.
%mem=256Mb
%chk=DCH3.chk
%nproc=1
#p HF/3-21G Freq=ReadIsotopes
DCH3
0 1
C
H 1 B1
H 1 B2 2 A1
H 1 B3 3 A2 2 D1
H 1 B4 3 A3 2 D2
B1 1.07000000
B2 1.07000000
B3 1.07000000
B4 1.07000000
A1 109.47120255
A2 109.47125080
A3 109.47121829
D1 -119.99998525
D2 120.00000060
blank line
350.0 1.0 temp pressure [scale]
12.0 isotope mass for atom 1
1.0 isotope mass for atom 2
1.0 isotope mass for atom 3
1.0 isotope mass for atom 4
2.0 isotope mass for atom 5
blank line
(5) Go to File and select Save. Close the Word Pad file. You will now be asked if you want to submit the job. Click OK.
(6) Once the job has finished, visualize the vibrational frequency and see how they have changed.
Manually editing files outside GaussView
You can edit the input files manually without using GaussView, either by opening the files in Word Pad or in Gaussian09W. Jobs can then be submitted directly using Gaussian09W.
Further information about Gaussian can be found at the Gaussian web site http://www.gaussian.com