Mod:Hunt Research Group/solvent
General
Quality of the approximation
Pay attention to the value of the density lying outside the cavity, i.e. inside the dielectric. In G03 this value is labeled as “error on total polarization charges”, and example from an output is given below. As a rule of thumb this value should be less than 0.05 for the calculation to be acceptable.
Error on total polarization charges = 0.03697
Introduction to the SMD Model
The SMD model is an implicit solvation model, which treats the solvent as a dielectric continuum. The solute is surrounded by a cavity, the walls of which can be thought of as representing the solvent. The cavity is built by first centering spheres on each solute atom and then combining these spheres. The calculated solvation energy (and the calculated wavefunction) is sensitive to the exact shape of this cavity.
Full details of how the SMD model works (and how it is parameterised) can be found in the original paper (doi: 10.1021/jp810292n). A second important paper (by the same authors) extends the model to ionic liquids, and includes parameters which should be used in the model for some example ionic liquids (doi: 10.1021/jp304365v).
The SMD model attempts to calculate both electrostatic (dependent on the solvent dielectric constant) and non-electrostatic (depends on a range of solvent descriptors, see below) contributions to solvation energy. The electrostatic solvation energy is directly dependent on the electron density of the solute, while the non-electrostatic solvation term depends solely on the solute geometry. The dependence of the electrostatic term on solute density means that using the SMD model will lead to changes in the wavefunction (relative to a gas phase calculation) as the electrostatic term must be taken into account during the scf procedure. If the geometry of a solute is fixed (e.g in a single-point energy calculation) then only the electrostatic terms will affect the calculated wavefunction.
NOTE: Two separate cavities are used in the SMD model, one for the electrostatic solvation energy, and one for the non-electrostatic solvation energy.
Solvent Descriptors
Different solvents are specified by using the experimental solvent descriptors below:
Static Dielectric Constant(ε) - Used for calculation of electrostatic terms. Accounts for solute-solvent electrostatic interactions. Higher values lead to more negative solvation energies. A list of static dielectric constants for common solvents can be found on the Gaussian website, under the SCRF keyword.
Refractive Index (n) - Used for calculation of non-electrostatic terms. The refractive index of a solvent is related to the strength of solute-solvent dispersion forces, higher values of n should lead to more negative solvation energies. Values can be found in the CRC handbook for chemistry (section 3 physical constants of organic compounds,http://www.hbcpnetbase.com/). WARNING its actually the refractive index squared which is entered into the gaussian input file (see below)
Abraham Hydrogen Bond Acidity (α) - Used for calculation of non-electrostatic terms. Also affects the shape of the electrostatic cavity if oxygen atoms are present in the solute, and can therefore affect the electrostatic part of the solvation energy. Used to account for solute-solvent hydrogen bonding where the solvent donates the hydrogen atom. Values for many solvents can be found in the supporting information of the original SMD paper (doi: 10.1021/jp810292n)
Abraham Hydrogen Bond Acidity (β) - Used for calculation of non-electrostatic terms. Used to account for solute-solvent hydrogen bonding where the solvent donates the lone pair. Values for many solvents can be found in the supporting information of the original SMD paper (doi: 10.1021/jp810292n)
Surface Tension (γ)- (Units of cal mol-1 Å-2)Used for calculation of non-electrostatic terms. Accounts for the energy required to create a cavity in the solvent (for the solute to fit into). Higher values of γ lead to more positive solvation energies (creating an empty cavity in a solvent is unfavorable). Values (in mN/m) can be found in the CRC handbook of chemistry (section 6-182, surface tensions of common liquids) but need to be multipled by 1.43932 to be converted to cal mol-1 Å-2.
Fraction of Solvent Atoms which are Aromatic Carbons (Φ) - Used for calculation of non-electrostatic terms. Accounts for systematic differences in solvation energies between aromatic/non-aromatic solvents. Can be worked out from the solvent chemical formula (hydrogen atoms are ignored). Examples of values for ionic liquids can be found in doi:10.1021/jp810292n
Fraction of Solvent Atoms which are Halogens (Ψ) - Used for calculation of non-electrostatic terms. Accounts for systematic differences in solvation energies between halogenated/non-halogenated solvents. Can be worked out from the solvent chemical formula (hydrogen atoms are ignored). Examples of values for ionic liquids can be found in doi:10.1021/jp810292n
Carrying Out Calculations With the SMD model
The keyword scrf (which stands for self-consistent reaction field) is used to carry out SMD calculated. The easiest way to do an SMD calculation is by using a pre-defined solvent in gaussian, in which case you just write scrf=(smd,solvent=Solv_Name) where the value of Solv_Name depends on which solvent you want to use. A list of pre-defined solvents is available on gaussians website, under the scrf keyword. For example heres an input file for an SMD calculation using pentane as the solvent.
%chk=Water_Molecule_SMD_Predef_pentane.chk # hf/3-21g scrf=(smd,solvent=n-pentane) Title Card Required 0 1 O -0.54187194 1.86371097 0.00000000 H 0.41812806 1.86371097 0.00000000 H -0.86232652 2.76864680 0.00000000
WARNING Gaussian will not print the parameters (except dielectric constant and refractive index) used to represent pre-defined solvents. Therefore you generally shouldn't use the pre-defined solvents.
Generally you should be defining the required solvent parameters yourself when carrying out an SMD calculation. To do this use the keywords scrf=(smd,solvent=generic) in the route line of the input file, and append a list of solvent descriptor values after the geometry/connectivity data. For example(again for the pentane solvent):
%chk=Water_Molecule_SMD_ReadIn_pentane_all.chk # hf/3-21g geom=connectivity scrf=(smd,solvent=generic) Title Card Required 0 1 O -0.54187194 1.86371097 0.00000000 H 0.41812806 1.86371097 0.00000000 H -0.86232652 2.76864680 0.00000000 1 2 1.0 3 1.0 2 3 eps=1.837100 epsinf=1.84 HBondAcidity=0.0 HBondBasicity=0.0 SurfaceTensionAtInterface=22.295067 CarbonAromaticity=0.0 ElectronegativeHalogenicity=0.0
Where:
eps = Dielectric Constant epsinf = refractive index SQUARED HBondAcidity = Abraham hydrogen bond acidity HBondBasicity = Abraham hydrogen bond basicity SurfaceTensionAtInterface = Surface Tenstion CarbonAromaticity = Fraction of Solvent Atoms which are Aromatic Carbons ElectronegativeHalogenicity = Fraction of Solvent Atoms which are Halogens
WARNING EPSINF=Refractive index SQUARED
Solvent parameters for example solvents
| Solvent | ε | n | α | β | γ | φ | Ψ |
|---|---|---|---|---|---|---|---|
| n-pentane | 1.8371 | 1.3575 | 0.0 | 0.0 | 22.30 | 0.0 | 0.0 |
| Decanol | 7.5305 | 1.4372 | 0.37 | 0.48 | 41.04 | 0.0 | 0.0 |
| [C4C1Im][SCN] | 13.7 | 1.54 | 0.18 | 0.52 | 68.29 | 0.231 | 0.0 |
| [C4C1Im][PF6] | 11.4 | 1.4090 | 0.266 | 0.216 | 70.24 | 0.1765 | 0.3529 |
Parameters for n-pentane, Decanol and [C4C1Im][PF6] are taken from the sources mentioned int he introduction. Parameters for [C4C1Im][SCN] are taken from: (doi: 10.1021/je101184s / doi: 10.1016/j.jct.2012.03.024 / doi: 10.1039/B9NJ00751B).
Important Information On the Implementation of the SMD model in Gaussian 09
The Treatment of Water
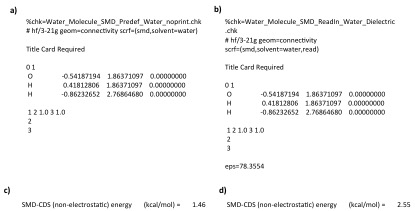
In the SMD model water is treated as a special solvent, in that the non-electrostatic terms are not actually calculated using solvent descriptors (unlike every other solvent). Therefore the correct way to carry out a calculation using the water solvent is to use the gaussian pre-defined solvent (scrf=smd,solvent=water). If water is defined in this way Gaussian will treat it as a special solvent. If ANY parameter is varied for the water solvent then Gaussian will NOT treat the solvent as water. For example, in the input file below (a) uses the pre-defined water solvent, while b) uses the same pre-defined solvent but reads in a dielectric constant (which is actually the one dielectric for the pre-defined water). These two input files lead to different non-electrostatic solvation energies being calculated despite the fact that both use the same solvent descriptors (panel c and d show non-electrostatic solvation energies calculated for input file a and b respectively). The reason is that when Gaussian read in the dielectric constant in b) it changed the solvent type from "water" to "generic" and therefore calculated the solvation energy in the same way it would for any other solvent.
NOTE: To generate the exact non-electrostatic solvation energy of input file b) by defining all terms yourself the following input file is used.
%chk=Water_Molecule_SMD_ReadIn_Water_GenericSolvent.chk # hf/3-21g geom=connectivity scrf=(smd,solvent=generic) Title Card Required 0 1 O -0.54187194 1.86371097 0.00000000 H 0.41812806 1.86371097 0.00000000 H -0.86232652 2.76864680 0.00000000 1 2 1.0 3 1.0 2 3 eps=78.3553 epsinf=1.777849 HBondAcidity=0.82 HBondBasicity=0.35 SurfaceTensionAtInterface=0.0 CarbonAromaticity=0.0 ElectronegativeHalogenicity=0.0
Meaning Gaussian 09 does not have a surface tension value for the pre-defined water solvent (103.63 cal mol-1 Å-2 is the surface tension of water).
The importance of using the "special" method of calculating non-electrostatic solvation energy for the water solvent, compared to reading in the correct parameters and treating it as a generic solvent, has been tested for multiple molecules (table below). Single point energy calculations were carried out for each structure (optimised at the HF/3-21 level in the gas phase) using both a user-defined water solvent (all relevant parameters read in, and the solvent treated as "generic") and the pre-defined Gaussian solvent (treated as a "special" solvent).
| Solute | Non-Electrostatic Solvation energy
(User Defined water, kJ mol-1) |
Non-Electrostatic Solvation energy
(Pre-defined water, kJ mol-1) |
User Defined - Pre-Defined
Non-Electrostatic Solvation energy (kJ mol-1) |
|---|---|---|---|
| [C4C1Im][SCN] ion pair | 40.1 | 29.3 | 10.8 |
| [C4C1Im]+ Cation | 31.5 | 24.3 | 7.2 |
| [SCN]- Anion | 20.7 | 14.3 | 6.4 |
| Methane | 13.2 | 11.4 | 1.8 |
| Water | 19.7 | 6.1 | 13.1 |
Kamlett Taft Vs Abraham Parameters
Values for Abraham hydrogen bond acidity and abraham hydrogen bond basicity are sometimes not available, while Kamlett-Taft values are (e.g the case for ionic liquids). As Abraham and Kamelett-Taft parameters both describe the strength of solute-solvent hydrogen bonds they can be related to each other. An empirical relation ship between Kamlett-Taft and Abraham hydrogen bonding parameters (see doi: 10.1021/jp304365v) is:
αAbraham = 0.4098 αKT + 0.0064
βAbraham = 0.6138 βKT + 0.0890
Where αAbraham and βAbraham are the Abraham hydrogen bond acidity and basicity respectively. αKT and βKT are the Kamlett-Taft hydrogen bond acidity and basicity respectively. Note that the relationship between Kamlet-Taft and Abraham parameters is not perfect, and it is therefore best to Abraham parameters which have been directly measured by experiment rather than converting Kamlet-Taft parameters to Abraham parameters.
Modifying the non-electrostatic Cavity
The cavity used to calculated electrostatic solvation energy can be modified using the normal cavity control keywords on the gaussian website (scrf keyword). The main reason i've done this is just to figure out what Gaussian 09 uses as defaults. Generally you want to use the default values, but there may be a situtation (although i cant think of one) where you'd want to modify the non-electrostatic cavity. To change the atomic radii used to construct the non-electrostatic cavity the keyword "NElRadii=" is used (and appended to the bottom of the input file. For example to use atomic radii from the UFF model the input file is:
%chk=Water_Molecule_SMD_ReadIn_NECavity_Testing3.chk # hf/3-21g geom=connectivity scrf=(smd,solvent=n-pentane) Title Card Required 0 1 O -0.54187194 1.86371097 0.00000000 H 0.41812806 1.86371097 0.00000000 H -0.86232652 2.76864680 0.00000000 1 2 1.0 3 1.0 2 3 NElRadii=UFF
Note: Gaussian seems to use Bondi Radii as default (as is suggested in the 2009 SMD paper) for the non-electrostatic cavity.
The non-electrostatic cavity is (by default, and as recommended in the 2009 SMD paper) a solvent accesible surface, meaning the the solvent radius is added to each of the atomic centered sphere when generating the cavity. For practical purposes this means that the shape of the non-electrostatic cavity depends on the solvent radius. The solvent radius used for the non-electrostatic cavity is modified with the keyword "SMDRSolvCDS=" using scrf=(smd,solvent=xxxxx,read) and inputting the value at the bottom of an input file. For example:
%chk=Water_Molecule_SMD_pentane_CDSRSolv_0pt3.chk # hf/3-21g geom=connectivity scrf=(smd,solvent=n-pentane,read) Title Card Required 0 1 O -0.54187194 1.86371097 0.00000000 H 0.41812806 1.86371097 0.00000000 H -0.86232652 2.76864680 0.00000000 1 2 1.0 3 1.0 2 3 SMDRSolvCDS=0.3
Note: Gaussian seems to use a non-electrostatic solvent radius of 0.4Å for all solvents (as is recommended in the 2009 SMD paper).