Mod:Hunt Research Group/Python scripts for cube files
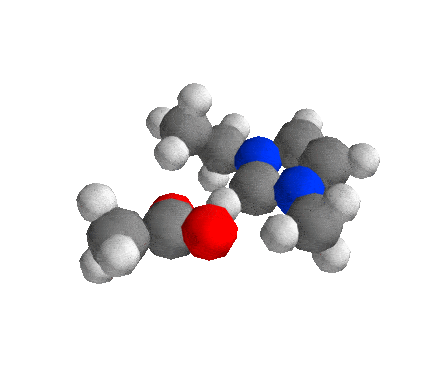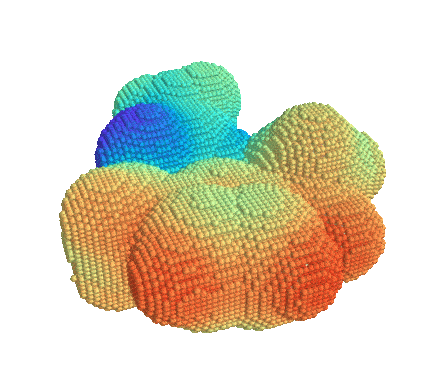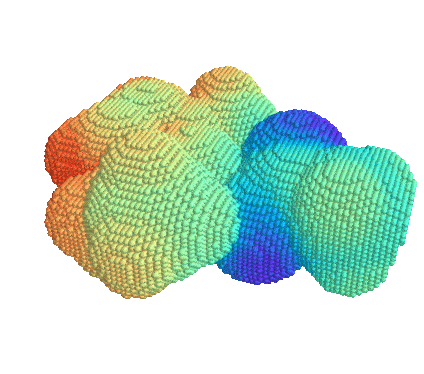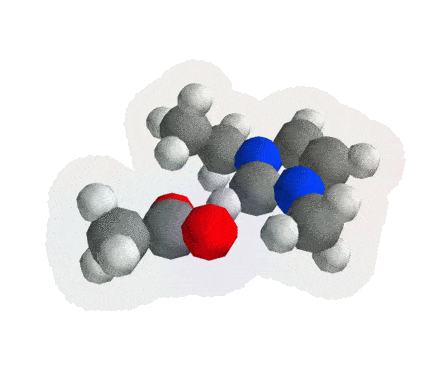
Getting Started
- go here for initial set-up getting started
The latest version of CubePy, along with a test cube file can be found here.
CubePy is under CC BY-NC-SA license
CubePy encompasses six modules that can be used to read, save, manipulate and visualise data from cube files:
- ReadCube.py - used to extract data from cube files and return it in convenient python formats
- SaveCube.py - used to save data from cube files in convenient formats
- CalculateCube.py - used to manipulate data from cube files
- PlotCube.py - used to plot data from cube files (using Matplotlib)
- PlotCube_mayavi.py - used to plot data from cube files (using Mayavi)
- AdditionalFunctions.py - used to manipulate data not from cube files, but from files produced by the scripts above
If you want to save data from cube files
- for saving the values from the cube files, use the functions from SaveCube.py
- for saving histogram data, use the function
PlotHistogramfrom PlotCube.py withsave=True - for saving a short summary file of the minimum, maximum and lowest curvature points on the van der Waals surface, use
save=Truewhen runningPlotSurfacefrom PlotCube_mayavi.py
If you want to plot surfaces, isosurfaces, slices
- use the functions from PlotCube_mayavi.py. They are faster and better at 3D plotting than the equivalent functions from PlotCube.py
If you want to generate static figures you can also consider using the 3D plotting functions from PlotCube.py. These are especially recommended when wanting to display atoms as they look better than their PlotCube_mayavi.py equivalents. However, keep in mind that Matplotlib sometimes fails at 3D rendering, which generates "incorrect" plots (i.e. elements that should be behind the rest of the plot appear at the front). Therefore, plots created using function from PlotCube.py should be double-checked (by comparing them with plots from PlotCube_mayavi.py, by example).
If you want to plot histograms and KDEs
- use
PlotHistogramfrom PlotCube.py to plot a histogram and/or KDE directly from cube files - use
OverlapKDEsandPlotHistogramfrom AdditionalFunctions.py to plot histograms and KDEs from files written byPlotHistogramfrom PlotCube.py whensave=True
