Mod:Hunt Research Group/Jan evaluate flex
Appearance
evaluating charges
editing the script
- the script we need is esp_fit_and_flex.py
- copy the script from repESP/scripts into your data directory, we will be editing it
- we need to know the number ordering of the atoms
- in our case the atoms are listed (by looking in the ch3oh_chelpg.esp file)
- 1 C
- 2, 3, 4 H (the CH3)
- 5 O
- 6 H (the OH)
- we will also need to manually evaluate equivalences
- atoms are now defined in order as a1, a2, a3 with equivalences defined
- so C is a1 with no equivalence
- in this molecule the H's 2, 3 and 4 are all equivalent so 2:a2 3:a2 4:a2
- the remaining atoms are not equivalent 5:a3 6:a4
- now set some specific items in the script
- esp_charge_type = 'mk' or to the MESH type you want to use
- alt_esp_charge_type = 'chelpg' or to the alternative charge you want to compare with NOTE this mesh needs to be in the same directory ie ch3oh_chelpg.esp
- molecule_name = 'ch3oh' this needs to be the name of the directory where files are stored
- vary_label1=1 atom number of atom to have its flexibility evaluated (methyl C in this case)
- charge_dict_1D = lambda a1: {1: a1} this C is not equivalent to any other atoms
- vary_label2 = 5 atom number of atom to vary along side the first atom to form a 2d graph (hydroxyl O in this case)
- charge_dict_2D = lambda a1, a3: {1:a1, 5:a3} and O is not equivalent to any other atoms
- labels_to_monitor = [5, 6, 2] we want to see how these other atoms vary as the C charge is altered
- xlim1 = (-0.5, 1) axis limits for the primary atom (C) this is a bit hit and miss so you may have to run several times
- xlim2 = (-1, 0) axis limits for the secondary atoms (H, O, H)
- sampling_num = 10 number of points to sample (start small and then increase, a number about 25 is good)
- swap_axes = False will swap axes if true
- plot_flexibility = True looks for single atom flexibility
- plot_response = True looks to find the response of a second atom (both of these should be true)
- thus the important bits for this example are
esp_charge_type = 'mk'
alt_esp_charge_type = 'chelpg'
molecule_name = 'ch3oh'
# Parameters to change when changing the molecule
# The first label is the more important atom, for which a 1D scan will be done
vary_label1 = 1
charge_dict_1D = lambda a1: {1: a1}
vary_label2 = 5
charge_dict_2D = lambda a1, a3: {1: a1, 5: a3}
# This shouldn't be more than 5 because that's the max number of colors in plot
labels_to_monitor = [5, 6, 2]
xlim1 = (-0.5, 1)
xlim2 = (-1, 0.5)
sampling_num = 21
swap_axes = False
plot_flexibility = True
plot_response = True
- if you want to tun off the charge variation figure (see below) and just do the two atom plot then
- look into the code for this and replace True with False
# ONE CHARGE VARIATION if True:
running the script
- to run the script we will use "python3.5 esp_fit_and_flex.py"
- this may take some time!
- a graph will appear you must manually save it and then close the graph for the code to continue!
- a new folder esp_fit_xx (where xx=charge method) will be created
- the same graph as above will appear as a pdf in this folder
- you will find the individual jobs in esp_fit_xx/resp_calcs
[tricia@dyn1242-77]/Users/tricia/bin/repESP/data $ python3.5 esp_fit_and_flex.py & [1] 15202 [tricia@dyn1242-77]/Users/tricia/bin/repESP/data $ esp_fit.py script Molecule: ch3oh Charge type: MK Non-ESP charge type: NBO Alt. ESP charge type: CHELPG Running unrestrained RESP to fit ESP with equivalence: Please check if the following RESP input is what you want: Atom 1: C Atom 2: H Atom 3: H , equivalenced to atom 2 Atom 4: H , equivalenced to atom 2 Atom 5: O Atom 6: H The molecule with MK charges: RMS: 0.00208 RRMS: 0.13573 RMSV: 0.01534 Atom 1: C , charge: 0.1747 Atom 2: H , charge: 0.0566 Atom 3: H , charge: -0.0084 Atom 4: H , charge: -0.0084 Atom 5: O , charge: -0.6056 Atom 6: H , charge: 0.3912 The molecule with equivalenced MK charges (unrestrained RESP): RMS: 0.00283 RRMS: 0.18461 RMSV: 0.01534 Atom 1: C , charge: 0.1237 Atom 2: H , charge: 0.0241 Atom 3: H , charge: 0.0241 Atom 4: H , charge: 0.0241 Atom 5: O , charge: -0.5448 Atom 6: H , charge: 0.3489 Checking differences between raw and equivalenced charges ... Atom 1: C differs by 0.051 Atom 5: O differs by -0.061
- so the first section generates MK charges
- then it works out the equivalenced charges
- and looks at the difference
- carrying on now ...
The molecule with equivalenced CHELPG charges (unrestrained RESP) evaluated on the MK grid: RMS: 0.00284 RRMS: 0.18542 RMSV: 0.01534 Percentage increase over equivalenced MK charges: 0.44 % Atom 1: C , charge: 0.1855 Atom 2: H , charge: 0.0076 Atom 3: H , charge: 0.0076 Atom 4: H , charge: 0.0076 Atom 5: O , charge: -0.5552 Atom 6: H , charge: 0.3470
- the secondary charges selection (ChelpG) is now evaluated on the MK grid
- carrying on some more ...
One-dimensional scan: Charges on these atoms will be varied: * Atom 1: C See below for equivalence information of other atoms. Please check if the following RESP input is what you want: Atom 1: C , frozen Atom 2: H Atom 3: H , equivalenced to atom 2 Atom 4: H , equivalenced to atom 2 Atom 5: O Atom 6: H Sampling progress: 100.00 % Flexibility limits on Atom 1: C 1% limits: 0.02654, 0.22079, diff: 0.19425 (157.1%) 5% limits: -0.09569, 0.34302, diff: 0.43871 (354.8%) 10% limits: -0.19032, 0.43765, diff: 0.62796 (507.8%) 20% limits: -0.33082, 0.57815, diff: 0.90897 (735.0%) Further limits were beyond the sampled range. The monitored charges at 10% flexibility limits: H2: 0.10642, -0.05831, diff: 0.16473 O5: -0.47959, -0.60996, diff: 0.13037 H6: 0.35064, 0.34725, diff: 0.00339
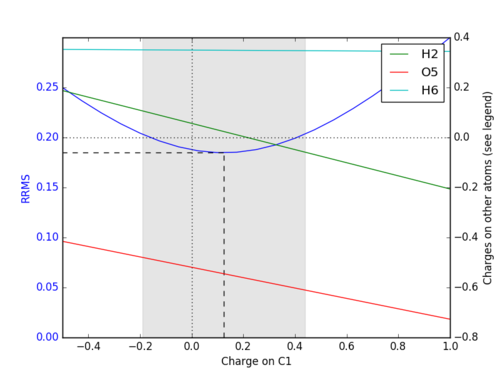
- this information relates to the graph produced
- example of the on-screen output for the first graph as shown
- left axis, is for the blue line and the dashed line
- for the primary atom (C in ch3oh in this case)
- dashed line gives minimum, minimum gives best fit to esp (+0.1855)
- flexibility limits are given as text on screen
- and the area greyed out is for a 10% variation in ESP
- this means the charge can be varied between these values with only a 10% degradation in the ESP or RRMS??
- right axis, is for the lightly dotted line, all other colours, and relates to the legend
- vertical line lightly dotted line shows zero charge
- horizontal lightly dotted relates to right axis where charges on other atoms are zero
- for example the green line is for H2, as the C becomes more positive the charge on H2 goes down
- at some point the H2 charge crosses the zero charge lightly dotted line, ie becomes negative
- at the min C charge H2 is just above zero
- the data files re in esp_fit_xx/resp_calcs in individual directories
- for example in our case the directories relate to varying the C atom charge
- each directory contains the run for a specific charge say +0.025 (a point on the blue line of the graph)
- reading the "punch" file gives relevant information (see below)
- the -1 under IVARY indicates that C is frozen at a charge of +0.025
- all other charges are then fitted taking into account equivalence
- for example the oxygen takes on the value of -0.52 which can be seen in the red line of the graph
Resp charges for organic molecule
iqopt irstrnt ihfree qwt
2 -1 1 0.000000
rel.rms dipole mom Qxx Qyy Qzz
0.18651 1.80453 1.49350 0.63919 -2.13269
Point charges before & after optimization
NO At.No. q0 q(opt) IVARY d(rstr)/dq
1 6 0.025000 0.025000 -1 0.000000
2 1 42.000000 0.049939 0 0.000000
3 1 42.000000 0.049939 2 0.000000
4 1 42.000000 0.049939 2 0.000000
5 8 42.000000 -0.524291 0 0.000000
6 1 42.000000 0.349474 0 0.000000
Statistics of the fitting:
The initial sum of squares (ssvpot) 0.099
The residual sum of squares (chipot) 0.003
The std err of estimate (sqrt(chipot/N)) 0.00286
ESP relative RMS (SQRT(chipot/ssvpot)) 0.18651
Dipole Moment (Debye)= 1.80453
evaluating the esp fit flexibility for two atoms
- this section takes much longer as there is more to compute
- the graph looks like this
- the cross in the center marks the MK charges in this case
- the plus marks the fitted ChelpG charge
- the filled circle is the NBO (NPA) charges
- the filled diamond is the point on the ESP surface with the same ratio as the NBO charges
- example of the on-screen output for the second graph as shown
Two-dimensional scan: Charges on these atoms will be varied: * Atom 1: C * Atom 5: O See below for equivalence information of other atoms. Please check if the following RESP input is what you want: Atom 1: C , frozen Atom 2: H Atom 3: H , equivalenced to atom 2 Atom 4: H , equivalenced to atom 2 Atom 5: O , frozen Atom 6: H Meshgrid progress: 100.00 % Scanning roughly various ratios. This shouldn't take long. Please check if the following RESP input is what you want: Atom 1: C , frozen Atom 2: H Atom 3: H , equivalenced to atom 2 Atom 4: H , equivalenced to atom 2 Atom 5: O , frozen Atom 6: H Starting minimization of charge ratio. FOUND optimal heavy ratio: 0.6904 with RRMS of 0.196694 Number of charges sampled along each axis: 21 Reporting contour surface areas: 1% contour area: 0.00025 5% contour area: 0.00470 10% contour area: 0.02172 20% contour area: 0.07508 30% contour area: 0.12503 50% contour area: 0.23843 100% contour area: 0.04076
- the data files are in esp_fit_xx/resp_calcs in individual directories
- for example in our case the directories relate to varying the C and O atom charges
- each directory contains the run for a specific C and O charge say C +0.025 and O -0.50
- the directory is called "C1+0.025-O5-0.050"
- reading the "punch" file gives relevant information (see below)
- the -1 under IVARY indicates that both the C and O are frozen at a charges of +0.025 and -0.50 respectively
- all other charges are then fitted taking into account equivalence
Resp charges for organic molecule
iqopt irstrnt ihfree qwt
2 -1 1 0.000000
rel.rms dipole mom Qxx Qyy Qzz
0.91587 0.18780 0.06899 0.05393 -0.12291
Point charges before & after optimization
NO At.No. q0 q(opt) IVARY d(rstr)/dq
1 6 0.025000 0.025000 -1 0.000000
2 1 42.000000 0.000804 0 0.000000
3 1 42.000000 0.000804 2 0.000000
4 1 42.000000 0.000804 2 0.000000
5 8 -0.050000 -0.050000 -1 0.000000
6 1 42.000000 0.022587 0 0.000000
Statistics of the fitting:
The initial sum of squares (ssvpot) 0.099
The residual sum of squares (chipot) 0.083
The std err of estimate (sqrt(chipot/N)) 0.01405
ESP relative RMS (SQRT(chipot/ssvpot)) 0.91587
Dipole Moment (Debye)= 0.18780