Mod1220
NH3 molecule
| N-H bond length | 1.01798 Å |
| H-N-H bond angle | 105.741 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -56.55776873 a.u. |
| RMS gradient | 0.00000485a.u. |
| Point Group | C3V |
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 1.018 -DE/DX = 0.0 ! ! R2 R(1,3) 1.018 -DE/DX = 0.0 ! ! R3 R(1,4) 1.018 -DE/DX = 0.0 ! ! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 ! ! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 ! ! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 ! ! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
NH3 molecule |
The optimisation file is liked to here
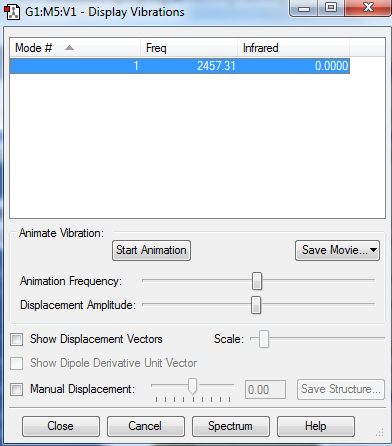
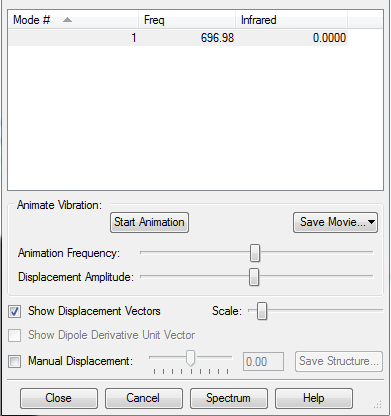
3 modes are expected from the 3N-6 rule.
Mode 2 and mode 3 are degenerate, mode 5 and mode 6 are degenerate.
Mode 1,2 and 3 are bending vibrations and mode 4,5,6 are bond stretch vibrations.
Mode 1 and 4 are highly symmetric.
Mode 1 is known as the umbrella mode.
Two bands are expected to be seen in a spectrum as mode 4,5,6 have very small intensities and mode 2 and 3 are degenerate.
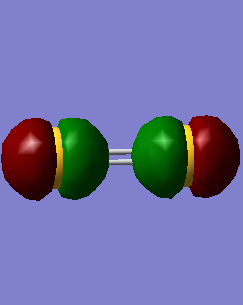
-1.125 charges on N , 0.375 charges on H.
N is expected to have negative charge as it is more electronegative than H, which means it has a stronger ability to attract electrons.
N2 molecule
| N-N bond length | 1.1055 Å |
| N-N bond angle | 90 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -109.52412868 a.u. |
| RMS gradient | 0.00000365a.u. |
| Point Group | D*H |
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000002 0.001800 YES RMS Displacement 0.000003 0.001200 YES ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
N2 molecule |
The optimisation file is liked to here
1 mode is expected from 3N-5 rule, which is stretch vibration.
It is highly symmetric.
Only one band is seen in spectrum.
Zero charge on both N atoms as there is no dipole in a diatomic molecule.
H2 Molecule
| H-H bond length | 0.74279 Å |
| H-H bond angle | 90 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -1.17853930 a.u. |
| RMS gradient | 0.00012170a.u. |
| Point Group | D*H |
Item Value Threshold Converged? Maximum Force 0.000211 0.000450 YES RMS Force 0.000211 0.000300 YES Maximum Displacement 0.000278 0.001800 YES RMS Displacement 0.000393 0.001200 YES ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 0.7431 -DE/DX = -0.0002 !
H2 molecule |
The optimisation file is liked to here
1 mode is expected from 3N-5 rule, which is stretch vibration.
It is highly symmetric.
Only one band is seen in spectrum.
Zero charge on both H atoms as there is no dipole in a diatomic molecule.
Reaction Energy of haber process
N2 + 3H2 -> 2NH3
| E(NH3) | -56.55776873 a.u. |
| 2*E(NH3 | -113.1155375 a.u. |
| E(N2) | -109.52412868 a.u. |
| E(H2) | -1.17853930 a.u. |
| 3*E(H2) | -3.5356179 a.u. |
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579092 a.u. = -146.479 kJ/mol
The ammonia product is more stable than the reactants as it has lower energy than reactants.
[| More information about Haber process]
Molecule S2
| S-S bond length | 1.92947 Å |
| S-S bond angle | 90 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -796.32599779a.u. |
| RMS gradient | 0.00000670 a.u. |
| Point Group | D*H |
Item Value Threshold Converged? Maximum Force 0.000012 0.000450 YES RMS Force 0.000012 0.000300 YES Maximum Displacement 0.000020 0.001800 YES RMS Displacement 0.000028 0.001200 YES ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 1.9295 -DE/DX = 0.0 !
S2 molecule |
The optimisation file is liked to here
1 mode is expected from 3N-5 rule, which is stretch vibration.
It is highly symmetric.
Only one band is seen in spectrum.
Zero charge on both S atoms as there is no dipole in a diatomic molecule.
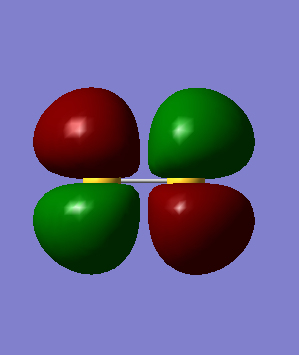
MOs of S2 molecule