MHZ15
Introduction
This is the page created by Hafiz Zainal for Computational Chemistry Lab 2. For parts 4 and 5, I have chosen the molecule chlorine monofluoride, ClF.
NH3 molecule
Key information

Calculation method: RB3LYP
Basis set: 6-31G(d.p)
E(RB3LYP): -56.55776873 a.u.
RMS gradient: 0.00000485 a.u.
Point group: C3V
N-H bond distance: 1.01798 Å
H-N-H bond angle: 105.741°
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986299D-10
Optimization completed.
-- Stationary point found.
NH3 molecule |
The optimisation file is linked to here.
Display vibrations
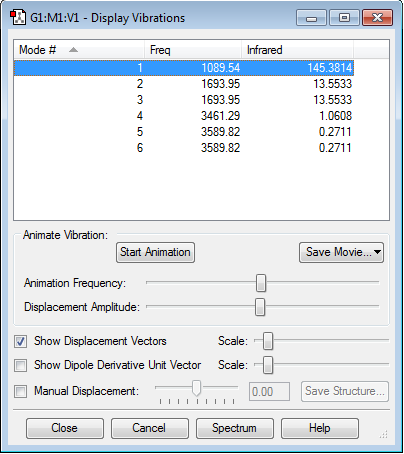
Q: How many modes do you expect from the 3N-6 rule?
A: There are 4 atoms, so N=4. 3N-6 rule gives 6 modes.
Q: Which modes are degenerate (ie have the same energy)?
A: Modes 2 and 3 are degenerate as well as 5 and 6.
Q: Which modes are "bending" vibrations and which are "bond stretch" vibrations?
A: Modes 1, 2 and 3 are "bending" vibrations while modes 4, 5 and 6 are "bond stretch" vibrations.
Q: Which mode is highly symmetric?
A: Mode 4 is highly symmetric.
Q: One mode is known as the "umbrella" mode, which one is this?
A: Mode 1 is the "umbrella" mode.
Q: How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
A: Two bands are expected because three (modes 1, 2 and 3) of the six modes have large enough absorption to be seen on the spectrum, but two out of those three are degenerate - they give the same peak. The reason modes 4, 5 and 6 have a very small absorption is that they are "stretching" modes, hence they have a considerably smaller change in dipole moment compared to that in "bending" modes of 1, 2 and 3.
Charge on the atoms
Charge on N-atom: 1.125e
Charge on H-atom: 0.375e
The expected charge on nitrogen is negative because it is more electronegative than hydrogen so it draws the bonding electrons more strongly. Hydrogen is expected to be positive because it is less electronegative so it draws the bonding electrons less strongly.
N2 molecule
Key information

Calculation method: RB3LYP
Basis set: 6-31G(d.p)
E(RB3LYP): -109.52412868 a.u.
RMS gradient: 0.00000060 a.u.
Point group: D∞h
N-N bond distance: 1.10550 Å
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400903D-13
Optimization completed.
-- Stationary point found.
N2 molecule |
The optimisation file is linked to here.
Display vibrations
H2 molecule
Key information

Calculation method: RB3LYP
Basis set: 6-31G(d.p)
E(RB3LYP): -1.17853936 a.u.
RMS gradient: 0.00000017 a.u.
Point group: D∞h
H-H bond distance: 0.74279 Å
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
H2 molecule |
The optimisation file is linked to here.
Display vibration
Reaction energy
Energies
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.1155375 a.u
E(N2)= -109.52412868 a.u
E(H2)= -1.17853936 a.u
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579082 a.u. = -146.48 kJ/mol
As the ΔE between the products and the reactants is negative, the ammonia product is more stable than the gaseous reactants.
ClF molecule
Key information
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
E(RB3LYP): -559.94269578 a.u.
RMS gradient: 0.00014211 a.u.
Point group: C∞V
Cl-F bond distance: 1.66434 Å
Item Value Threshold Converged?
Maximum Force 0.000246 0.000450 YES
RMS Force 0.000246 0.000300 YES
Maximum Displacement 0.000433 0.001800 YES
RMS Displacement 0.000613 0.001200 YES
Predicted change in Energy=-1.066054D-07
Optimization completed.
-- Stationary point found.
ClF molecule |
The optimisation file is linked to here.
Display vibration
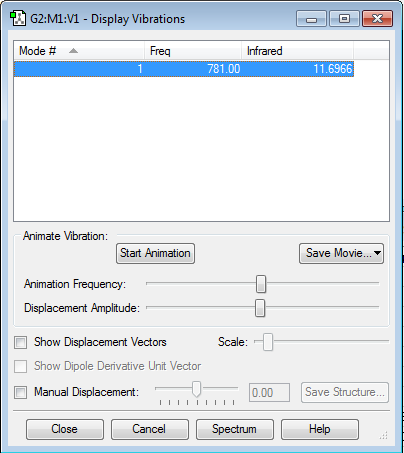
Based on the 3N-5 rule for linear molecules, 1 mode of vibration is expected. There should be one peak in the fingerprint region corresponding to frequency 781Hz visible on an IR spectrum of ClF due to the absorption being large enough for it to be seen.
Charge distribution
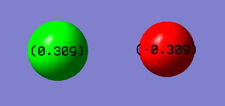
The charge on both Cl and F atoms is the same - 0.309. The charge on the F-atom, however, is negative and that on the Cl-atom is positive, cancelling out the charges and making the molecule a neutral one.
Molecular orbitals
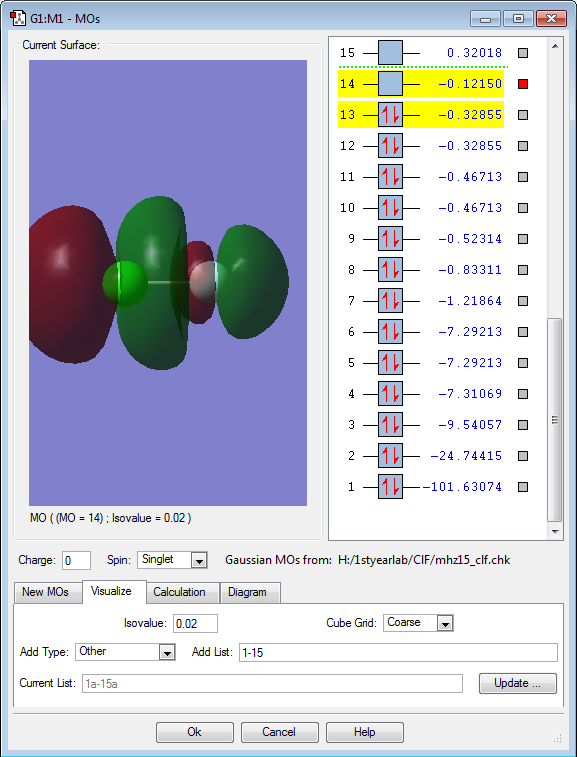
This molecular orbital is the lowest unoccupied molecular orbital, and in the case of ClF, this is the anti-bonding 4σ orbital with 3 nodes. This MO is very high in energy.
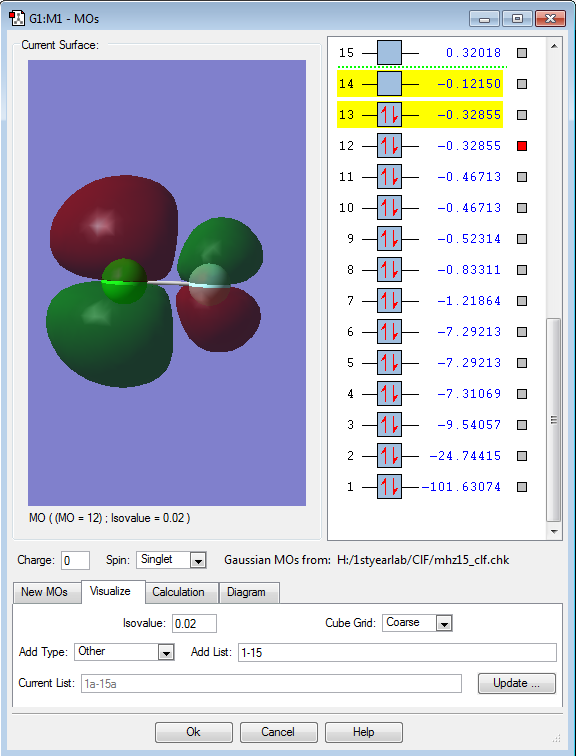
Molecular orbitals 12 and 13 are on the same energy level - they are degenerate. From the image alone we can see that it has 2 nodes, and since it is the highest occupied molecular orbital in ClF, it is obvious that this is the bonding 2π orbital. This is the result of combining 3p-electrons from Cl and 2p-electrons from F. MOs 12 and 13 are slightly higher in energy than the 3p-orbital of Cl, which is the HOMO in the case of ClF.
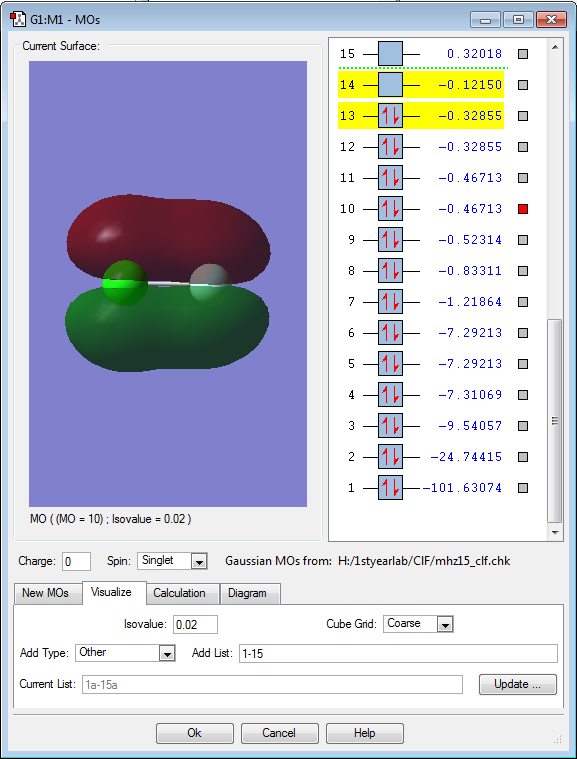
Molecular orbitals 10 and 11 are also degenerate, and both orbitals have 1 node each. Because they are degenerate and have 1 node, we can infer that they are the bonding 1π orbital, that result from combining 3p-electrons from Cl and 2p-electrons from F. MOs 10 and 11 are in the region of the HOMO/LUMO (3p orbitals in Cl and 2p orbitals in F, respectively).
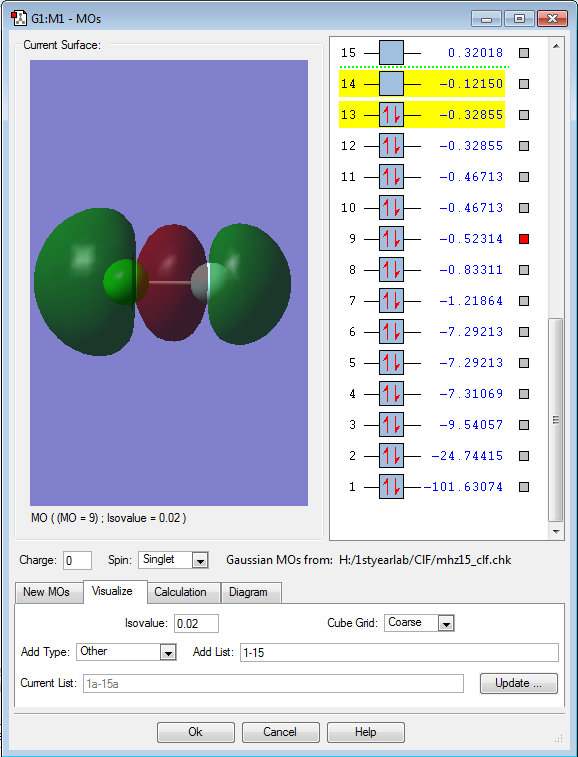
Molecular orbital 9 is the highest non-degenerate occupied molecular orbital. It has 2 nodes, thus making it obvious that it is the bonding 3σ orbital, formed by an electron from 3p-orbital of Cl and an electron from 2p-orbital of F.
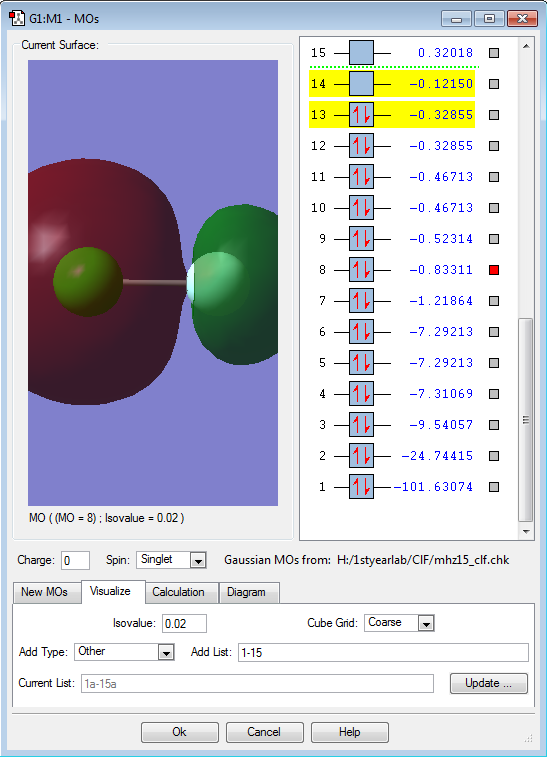
Molecular orbital 8 is the second highest non-degenerate occupied molecular orbital. It has one node. It is the anti-bonding 2σ orbital that results from combining a 3s-electron in Cl and a 2s-electron in F. MO 8 is in the region of the HOMO/LUMO (3s orbital in Cl and 2s orbital in F, respectively).
