Lo915
NH3 Molecule
- Calculation method: RB3LYP
- Basis set: 6-31G(d,p)
- Final energy: -56.55776873 a.u.
- RMS gradient: 0.00000485 a.u.
- Point group: C3v
- N-H optimised bond length: 1.01798 Å
- H-N-H optimised bond angle: 105.741°
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986272D-10 Optimization completed.
NH3 molecule |
The optimisation file is linked to here
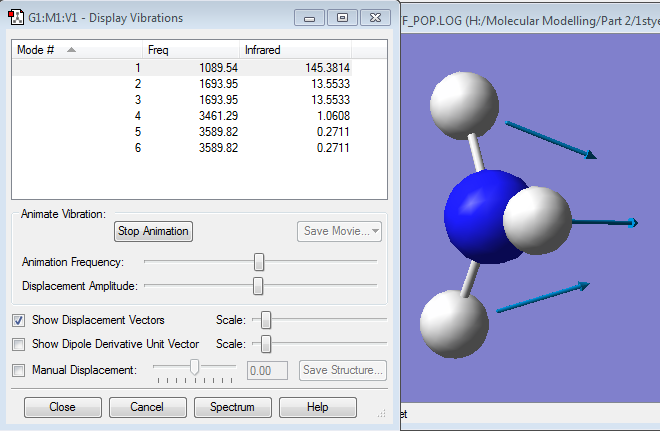
Vibrations
From the 3N-6 rule, there are 6 vibrational modes. Modes 2 and 3, and modes 5 and 6 are degenerate. Modes 1, 2 and 3 are "bending vibrations", while modes 4, 5 and 6 are "bond stretches". Mode 4 is highly symmetric. Mode 1 is known as the "umbrella" mode. You would expect to 2 bands see in an experimental spectrum of gaseous ammonia as only 3 of the modes have a high enough intensity in the infrared, and 2 of these are degenerate so only 1 peak would show for these 2 modes.
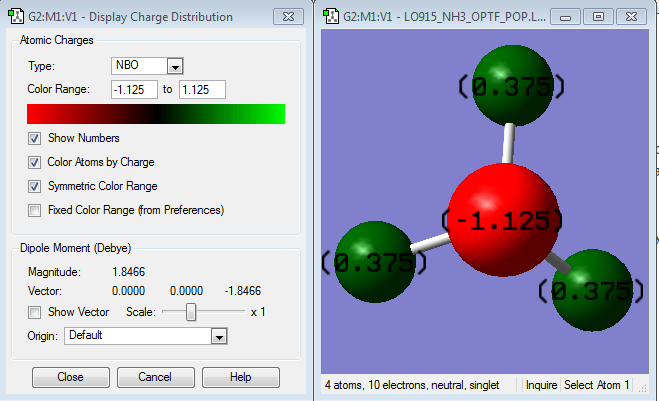
Charge distribution
The charge on the nitrogen is -1.125 and the charge on each of the hydrogens is +0.375. This is as expected as nitrogen is more electronegative, so would draw electron density towards itself, away from the hydrogens, causing the nitrogen to be negative and leaving the hydrogens positive
N2 Molecule
- Calculation method: RB3LYP
- Basis set: 6-31G(d,p)
- Final energy: -109.52412868 a.u.
- RMS gradient: 0.0000006 a.u.
- Point group: D∞H
- N-N optimised bond length: 1.10550 Å
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.400905D-13 Optimization completed.
N2 molecule |
The optimisation file is linked to here
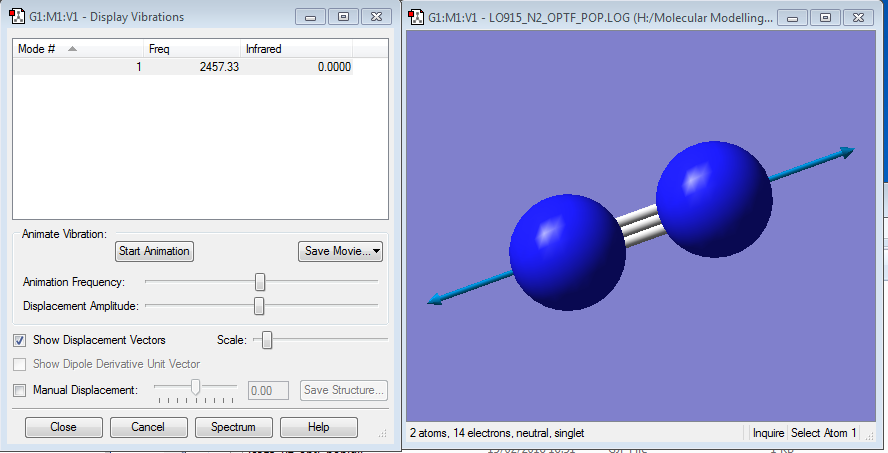
Vibrations
There is only one vibration as there is only one bond in this molecule, there would be no peak in an IR spectrum as there is no dipole in N2
H2 Molecule
- Calculation method: RB3LYP
- Basis set: 6-31G(d,p)
- Final energy: -1.17853935 a.u.
- RMS gradient: 0.00005224 a.u.
- Point group: D∞H
- H-H optimised bond length: 0.74292 Å
Item Value Threshold Converged? Maximum Force 0.000090 0.000450 YES RMS Force 0.000090 0.000300 YES Maximum Displacement 0.000119 0.001800 YES RMS Displacement 0.000168 0.001200 YES Predicted change in Energy=-1.077361D-08 Optimization completed.
H2 molecule |
The optimisation file is linked to here
Vibrations
There is only one vibration as there is only one bond in this molecule, there would be no peak in an IR spectrum as there is no dipole in H2
Haber-Bosch reaction
N2 + 3H2 → 2NH3
- E(NH3)= -56.55776873 a.u.
- 2*E(NH3)= -113.11553746 a.u.
- E(N2)= -109.52412868 a.u.
- E(H2)= -1.17853935 a.u.
- 3*E(H2)= -3.53561805 a.u.
- ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579073 a.u. = -146.48 kJ/mol
The product (ammonia) is more stable than the gaseous products as the reaction is exothermic (negative reaction energy) , so the product is at a lower energy than the reactants
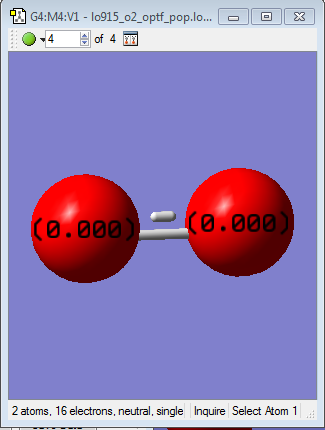
O2 (Project molecule)
- Calculation method: RB3LYP
- Basis set: 6-31G(d,p)
- Final energy: -150.32004019 a.u.
- RMS gradient: 0.00006587a.u.
- Point group: D∞H
- O-O optimised bond length: 1.21461Å
Item Value Threshold Converged? Maximum Force 0.000114 0.000450 YES RMS Force 0.000114 0.000300 YES Maximum Displacement 0.000069 0.001800 YES RMS Displacement 0.000097 0.001200 YES Predicted change in Energy=-7.817745D-09 Optimization completed.
O2 molecule |
The optimisation file is linked to here
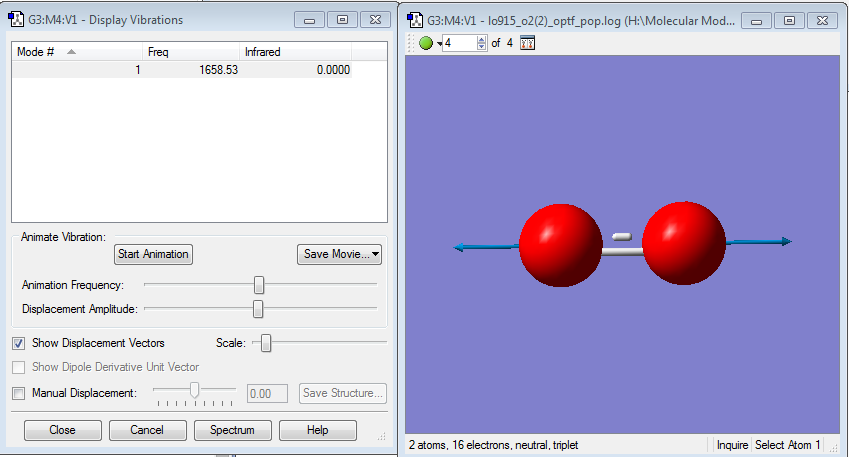
Vibrations
There is only one bond in this molecule, so the only vibration is the stretching of this bond. As this is a diatomic molecule, there are no bending vibrations. There would be no peak in an IR spectrum as the molecule so not have a dipole, so there is no change in dipole with the vibration.
Charge distribution
O2 is a homonuclear molecule, so there is no difference in electronegativity between the individual atoms as they are the same. This means that the electrons are distributed evenly over the molecule, so there is no charge on either atom, as seen below.
Molecular Orbitals
Chloroethane
- Calculation method: RB3LYP
- Basis set: 6-31G(d,p)
- Final energy: -539.43303632a.u.
- RMS gradient: 0.00008597a.u.
- Point group: CS
Item Value Threshold Converged? Maximum Force 0.000130 0.000450 YES RMS Force 0.000046 0.000300 YES Maximum Displacement 0.001070 0.001800 YES RMS Displacement 0.000409 0.001200 YES Predicted change in Energy=-1.847136D-07 Optimization completed.
CH3CH2Cl |
The optimisation file is linked to here
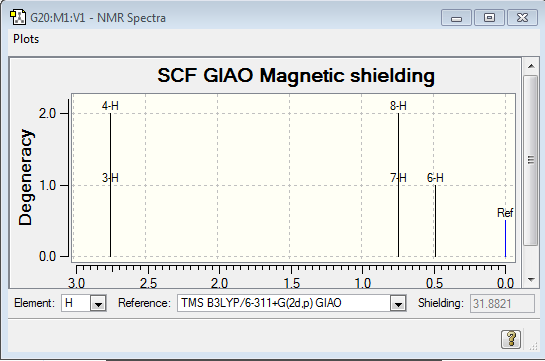
NMR: This is the NMR spectra calculated by Gaussview:
This does not match the expected NMR, which is confirmed when analysed experimentally, as shown here:
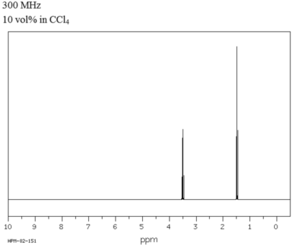
This shows that the calculated values have a slightly lower shift than the actual values, and in the calculated spectrum, the hydrogens from CH3 have been split into two environments, with the relative intensity of 2:1