LL:IM2
General aims
Identify the structure of NH3, H2 and N2
Identify the Haber-Bosch reaction energy calculation
NH3
General View of Molecular Structure
| Parameter | Result |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d.p) |
| final energy E | -56.55776873 a.u. |
| RMS gradient | 0.00000485 a.u. |
| point group | C3V |
| Bond Angle | 105.741 Degrees |
| Bond Length | 1.01798 Angstroms |
Forces and Displacement
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986286D-10
Image of Sturcture
NH3 |
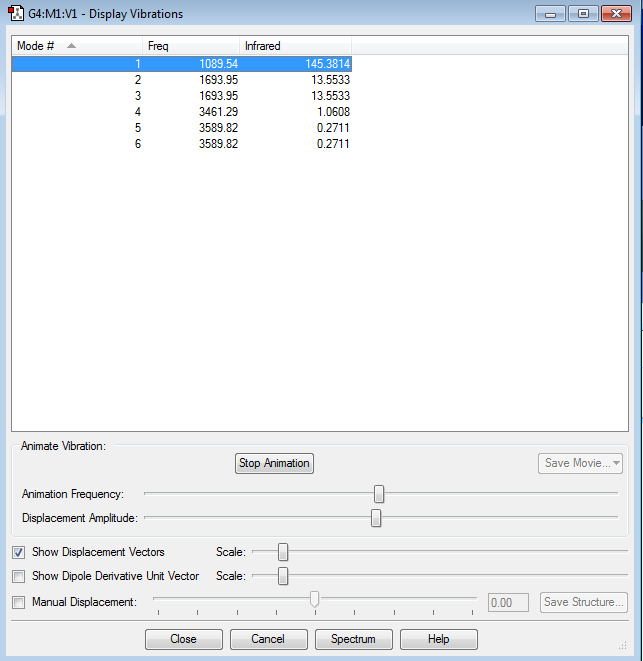
Vibration of Molecule
Display of Vibration
Data Included From the Vibration Data
how many modes do you expect from the 3N-6 rule?
Six Modes.
which modes are degenerate (ie have the same energy)?
Mode 2&3, Mode 5&6
which modes are "bending" vibrations and which are "bond stretch" vibrations?
Mode 1,2 &3 are bending vibrations. Mode 4,5 &6 are bond stretch vibrations.
which mode is highly symmetric?
Mode 4
one mode is known as the "umbrella" mode, which one is this?
Mode 1
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
Two bands in total because mode 2&3 are degenerate orbitals which has the same wavelength, thus two bands merged. For mode 4,5&6, the intensity is approximate to 0, which is too small to be observed in the spectrum. So overall, two bands shown in the spectrum.
Bond Charge
Charge on the N-atom= -1.125 c Charge on the H-atom= 0.375 c Because the electronegativity of nitrogen is stronger than hydrogen.
N2
General View of Molecular Structure
| Parameter | Result |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d.p) |
| final energy E | -109.52412867 a.u. |
| RMS gradient | 0.00004425 a.u. |
| point group | D*H |
| Bond Angle | 180.00 Degrees |
| Bond Length | 1.10552 Angstroms |
Forces and Displacement
Item Value Threshold Converged? Maximum Force 0.000077 0.000450 YES RMS Force 0.000077 0.000300 YES Maximum Displacement 0.000024 0.001800 YES RMS Displacement 0.000034 0.001200 YES Predicted change in Energy=-1.836250D-09
Image of Sturcture
N2 |
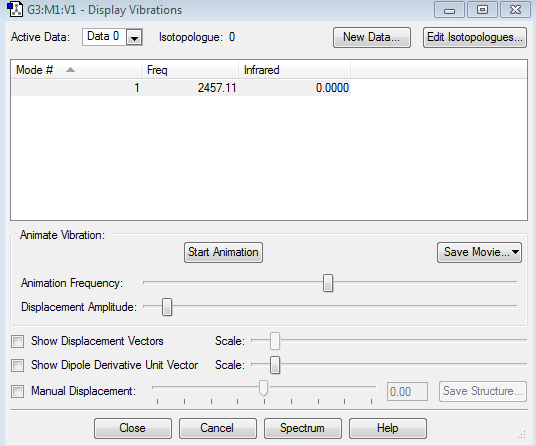
Vibration of Molecule
Display of Vibration
Data Included From the Image
how many modes do you expect from the 3N-5 rule?
one mode, the structure is non-linear, follows 3N-5 rule, thus only gives one mode.
Bond Charge
Charge on the N-atom= 0.000 c Charge on the N-atom= 0.000 c N atoms has no overall charge because the the molecule is non polar.
H2
General View of Molecular Structure
| Parameter | Result |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d.p) |
| final energy E | -1.17853930 a.u. |
| RMS gradient | 0.00011791 a.u. |
| point group | D*H |
| Bond Angle | 180.00 Degrees |
| Bond Length | 0.74308 Angstroms |
Forces and Displacement
Item Value Threshold Converged? Maximum Force 0.000204 0.000450 YES RMS Force 0.000204 0.000300 YES Maximum Displacement 0.000269 0.001800 YES RMS Displacement 0.000380 0.001200 YES Predicted change in Energy=-5.493616D-08
Image of Sturcture
H2 |
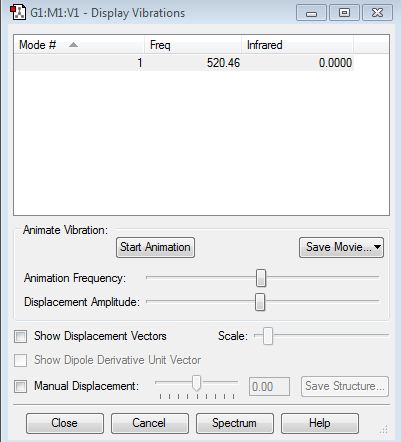
Vibration of Molecule
Display of Vibration
Data Included From the Virbration
how many modes do you expect from the 3N-5 rule?
one mode
Bond Charge
Charge on the N-atom= 0.000 c Charge on the H-atom= 0.000 c Because the electronegativity two hydrogen atoms are the same .
energy for the reaction of N2 + 3H2 -> 2NH3
E(NH3)=-56.56 a.u.
2*E(NH3)=-113.12 a.u.
E(N2)=-109.52 a.u.
E(H2)=-1.18 a.u.
3*E(H2)=-3.54 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05579093 a.u.=-146.48 KJ/mol
All data approximated to 2d.p.
compare with literature
The change in energy of Haber process is -92.4 KJ/mol. The literature value is lower then the calculation. That is because the optimization is not finished. Higher amount of energy released is calculated.
Stability of Reaction
Product more stable because the total energy of reaction is negative, energy produced in the forward reaction. The total energy of the product is smaller than the total energy of the reactant, thus the ammonia product is more stable.
Cl2
General View of Molecular Structure
| Parameter | Result |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d.p) |
| final energy E | -920.34987887 a.u. |
| RMS gradient | 0.00000468 a.u. |
| point group | D*H |
| Bond Angle | 180.000 Degrees |
| Bond Length | 2.04163 Angstroms |
Forces and Displacement
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000008 0.000300 YES Maximum Displacement 0.000023 0.001800 YES RMS Displacement 0.000032 0.001200 YES Predicted change in Energy=-1.829341D-10
Image of Sturcture
NH3 |
Vibration of Molecule
Display of Vibration
Data Included From the Vibration Data
how many modes do you expect from the 3N-5 rule?
One Mode. Because the molecule is non-polar, follows the 3N-5 rule. Therefore there is only one mode.
Bond Charge
Charge on the Cl-atom= 0.000 c Because the electronegativity of chlorine molecule is the same. The molecule itself has no charge, as it is non-polar.
Molecular Orbital
Chlorine atom with 17 electrons in each atom, with the structure of 1s22s22p63s23p5. Cl2 froms 17 molecular orbitals in total.
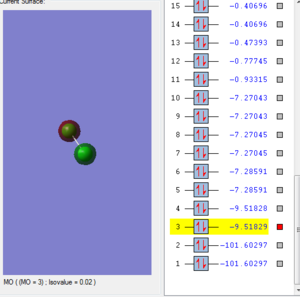
This molecular orbital is consisted by two 2s atomic orbital from each chlorine atom, which are in different phase. The shape of the structure is not symmetrical. There are two electrons in this molecular orbital. The orbital is anti-bonding since two AOs are in different phase. The energy of 2s bonding orbital is -9.51829 KJ/mol. It is very low in energy so that it is not likely to break bond in the reaction.
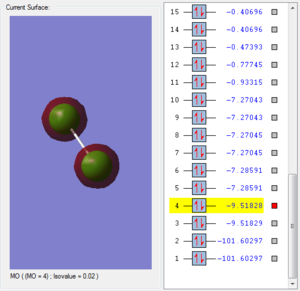
This molecular orbital is consisted of two 2s atomic orbitals from two chlorine atoms. The structure of the orbital is symmetric because two atomic orbitals have the same phase. Thus the structure is symmetric. The energy of the 2s anti-bonding orbtial is about same to the 2s bonding orbital.
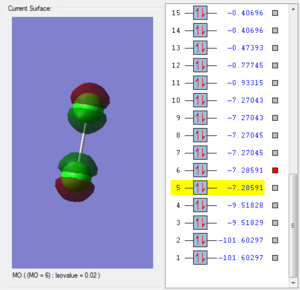
This molecular orbital is consisted by two 2px each from one chlorine atom, which are in phase with one another. There are two electrons in this MO. The energy of 2px bonding orbital is lower then the energy of 2py because the energy of 2p orbitals are not degenerate. 2px orbital and 2s orbital are in the same symmetry, thus they undergo mixing. So the energy of 2px orbital is smaller than 2py orbital. It is less likely to break in the reaction than the 2py molecular orbital. But still, both of the orbitals are low in energy and not likely to break during in the reaction.
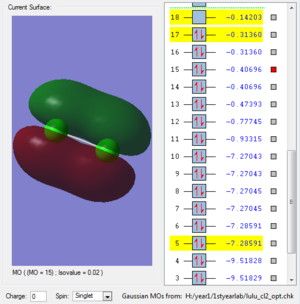
This molecular orbital is formed by two 3py atomic orbitals overlap horizontally with one another. A sigma bond is formed.Two 3p atomic orbitals are in different phase. The orbital is anti-bonding. There are two electrons in this molecular orbital. The structure is not symmetrical. This molecular orbital has higher energy so it is more likely to break in the reaction. It will break when 17th molecular orbital breaks.
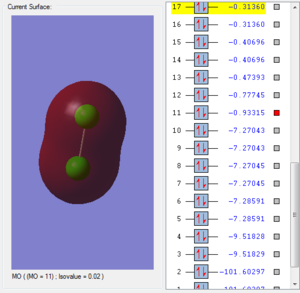
This molecular orbital is consisted of two 3s atomic orbitals from each chlorine atom. This orbital is occupied by two electrons. Two atomic orbitals are in the same phase. This orbital is symmetrical. The energy of 3s molecular orbital is the higher than the 2s molecular orbital and the size of the molecular orbital is bigger because electron in 3s orbital experiences larger degree of shielding so the attractive froce between the nucleus and the electrons is smaller, electrons are more diffused. The energy of 3d molecular orbital is higher make this bond more likely to break in the reaction. The energy of this molecular orbital is not the highest.