KSM214
NH3 molecule
The following information summarizes the the NH3 molecule that I have optimized using the Gaussian tool.
Analysis
| Molecule | NH3 |
| Calculation method | FREQ |
| Basis set | 6-1G(D.P) |
| Final energy | -56.55776873 a.u. |
| RMS gradient | 0.00000485 a.u. |
| Point group | C3v |
| Bond length | 1.01798 Å |
| Bond angle | 105.74118° |
| Item | Value | Threshold | Converged? |
|---|---|---|---|
| Maximum Force | 0.000004 | 0.000450 | YES |
| RMS Force | 0.000004 | 0.000300 | YES |
| Maximum Displacement | 0.000072 | 0.001800 | YES |
| RMS Displacement | 0.000035 | 0.001200 | YES |
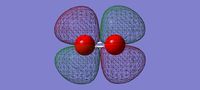
3D image of the NH3 Molecule
NH Molecule |
The optimisation file has been linked here
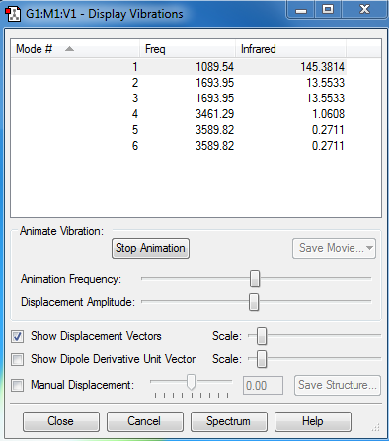
Molecular Vibrations
| Number of modes to be expected: | 6 |
| Degenerate modes: | 2,3 and 5,6 |
| Bending vibrations: | 1,2,3 |
| Stretching vibrations: | 4,5,6 |
| Symmetric mode: | 4 |
| Umbrella mode: | 1 |
| Bands expected: | 2 |
| Charge on Nitrogen atom: | -1.25 |
| Charge on Hydrogen atom: | 0.375 |
N2 Molecule
The following information summarizes the the N2 molecule that I have optimized using the Gaussian tool.
Analysis
| Molecule | N2 |
| Calculation method | RB3LYP |
| Basis set | 6-3-1G(d.p) |
| Final energy | -109.52412868 a.u. |
| RMS gradient | 0.00000060 a.u. |
| Point group | D*H |
| Bond length | 1.10550 Å |
| Bond angle | 180° |
| Item | Value | Threshold | Converged? |
|---|---|---|---|
| Maximum Force | 0.000001 | 0.000450 | YES |
| RMS Force | 0.000001 | 0.000300 | YES |
| Maximum Displacement | 0.000000 | 0.001800 | YES |
| RMS Displacement | 0.000000 | 0.001200 | YES |
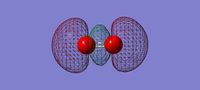
3D image of the N2 molecule
N Molecule |
The optimisation file has been linked here
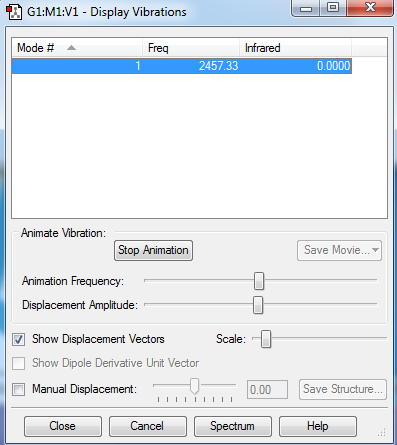
Molecular Vibrations
The N2 molecule being a diatomic molecule has a charge equal to zero.
H2 Molecule
The following information summarizes the the N2 molecule that I have optimized using the Gaussian tool.
Analysis
| Molecule | H2 |
| Calculation method | RB3LYP |
| Basis set | 6-3-1G(d.p) |
| Final energy | -1.17853936 a.u. |
| RMS gradient | 0.00000017 a.u. |
| Point group | D*H |
| Bond length | 0.74279 Å |
| Bond angle | 180° |
| Item | Value | Threshold | Converged? |
|---|---|---|---|
| Maximum Force | 0.000000 | 0.000450 | YES |
| RMS Force | 0.000000 | 0.000300 | YES |
| Maximum Displacement | 0.000000 | 0.001800 | YES |
| RMS Displacement | 0.000001 | 0.001200 | YES |
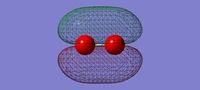
3D image of the H2 molecule
H Molecule |
The optimisation file has been linked here
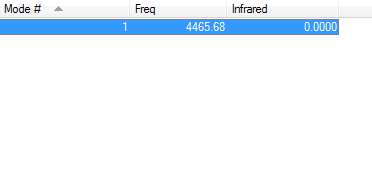
Molecular Vibrations
The H2 molecule being a diatomic molecule has a charge equal to zero.
Energy Calculations for the Haber-Bosch process
The Haber-Bosch process is a reaction in which nitrogen gas and hydrogen gas are converted to ammonia. It is an extremely important reaction within the farming industry as a fertilizer.
N2(g) + 3H2(g) → 2NH3(g)
The Enthalpy of the reaction has been calculated below using the table of results that I have acquired from the summary data of the optimized molecules.
| E(NH3) | -56.55776873 a.u. |
| 2*E(NH3) | -113.1155374 a.u. |
| E(N2) | -109.52412868 a.u. |
| E(H2) | -1.17853936 a.u. |
| 3*E(H2) | -3.53561808 a.u. |
| ΔE=2*E(NH3)-[E(N2)+3*E(H2)] | -0.05578994 |
| ΔE | -146.476 kj/mol |
O2 Molecule
The following information summarizes the the O2 molecule that I have optimized using the Gaussian tool.
Analysis
| Molecule | H2 |
| Calculation method | FREQ |
| Basis set | 6-3-1G(D.P) |
| Final energy | -150.25742434 a.u. |
| RMS gradient | 0.00007502 a.u. |
| Point group | D*H |
| Bond length | 1.21602 Å |
| Bond angle | 180° |
| Item | Value | Threshold | Converged? |
|---|---|---|---|
| Maximum Force | 0.000130 | 0.000450 | YES |
| RMS Force | 0.000130 | 0.000300 | YES |
| Maximum Displacement | 0.000080 | 0.001800 | YES |
| RMS Displacement | 0.000113 | 0.001200 | YES |
3D image of the O2 Molecule
O Molecule |
The optimisation file has been linked here
Molecular Vibrations
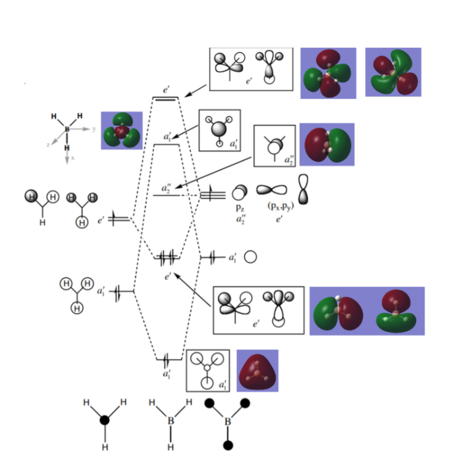
Molecular Orbitals