Jxpage1
NH3 Molecule Investigation
Optimization Parameters
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -56.55776873 a.u.
RMS Gradient Norm: 0.00000485 a.u.
Point Group: C3v
Calculated Parameters
N-H bond length = 1.01798 Å
H-N-H bond angle = 105.741°
NH3 Molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Vibration Analysis
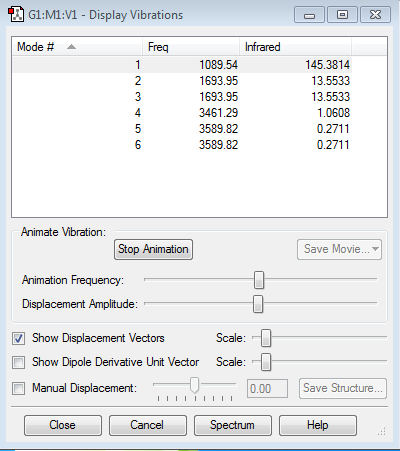
Number of modes expected: 6 (according to 3N-6 rule and 3*4-6=6)
Degenerated modes: a) Mode #2 & #3, and b) Mode #5 & #6
Bending vibrations: #1, #2 and #3
Stretching vibrations: #4. #5 and #6
Highly symmetric: #4
"Umbrella" mode: #1
Number of bands expected in an spectrum: 2 (because the #4, #5 and #6 have too small values to be shown on a spectrum)
Charge Distribution
N atom: -1.125
H atom: +0.375
Expectation: The N atom should have a partly negative charge and H atoms should have partly positive charge. Due to higher electronegativity of nitrogen, it attracts electrons more. Therefore, the N-H bond is polarised, and electrons are closer to N atom, which causes a partly negative charge on it. The calculation fits the expectation well.
H2 Molecule Investigation
Optimization Parameters
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -1.17853936 a.u.
RMS Gradient Norm: 0.05375030 a.u.
Point Group: Dinfh
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
The optimisation file is liked to here
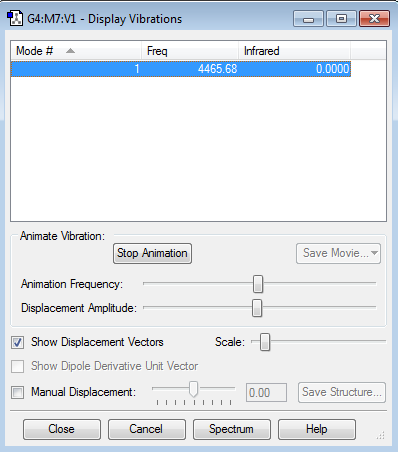
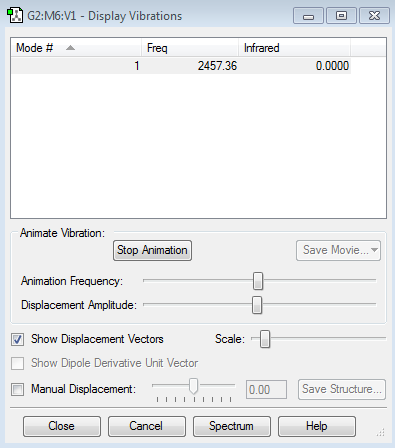
Vibrations:
N2 Molecule Investigation
Optimization Parameters
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -109.52412868 a.u.
RMS Gradient Norm: 0.18673398 a.u.
Point Group: Dinfh
Item Value Threshold Converged?
Maximum Force 0.000010 0.000450 YES
RMS Force 0.000010 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000004 0.001200 YES
The optimisation file is liked to here
Vibrations:
NH3 Energy calculations
E(NH3)= -56.55776873 a.u.
2E(NH3)= -113.11553746 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853936 a.u.
3E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05576070 a.u.=-146.40 kJ/mol
Due to negative ΔE, the product is more stable.
PH5 Molecule Investigation
Optimization Parameters
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -344.25491049 a.u.
RMS Gradient Norm: 0.00000471 a.u.
Point Group: D3h
Calculated Parameters
P-H bond length = 1.48687 Å (axial) and 1.43316 Å (equatorial)
H-P-H bond angle = 120°(equatorial-equatorial), 90°(equatorial-axial), 180°(axial-axial)
PH5 Molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000009 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000055 0.001800 YES
RMS Displacement 0.000022 0.001200 YES
Vibration Analysis
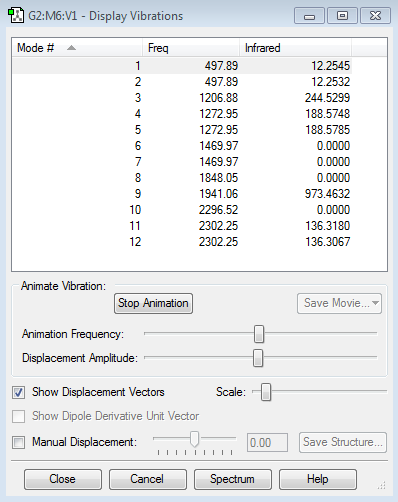
Number of modes expected: 12 (according to 3N-6 rule and 3*6-6=6)
Degenerated modes: a) Mode #1 & #2, b) #4 & #5, c) #6 & #7, d) #11 & #12
Bending vibrations: #1 to #7
Stretching vibrations: #8 to #12
Highly symmetric: #10
Number of bands expected in an spectrum: 5 (because #6 to #8 and #10 have zero values so that they cannot be shown on a spectrum)
Charge Distribution
P atom: +0.412
H atom: -0.180
Expectation: The H atoms should have a partly negative charge and the P atom should have partly positive charge, but the difference should be small. Due to slightly higher electronegativity of hydrogen (2.20 for H and 2.19 for P), it attracts electrons a little more. Therefore, the P-H bond is polarised, and electrons are closer to H atoms, which causes a partly negative charge on them. However, the difference in electronegetivity is so small that values for charges should be small. The calculation fits the expectation well.
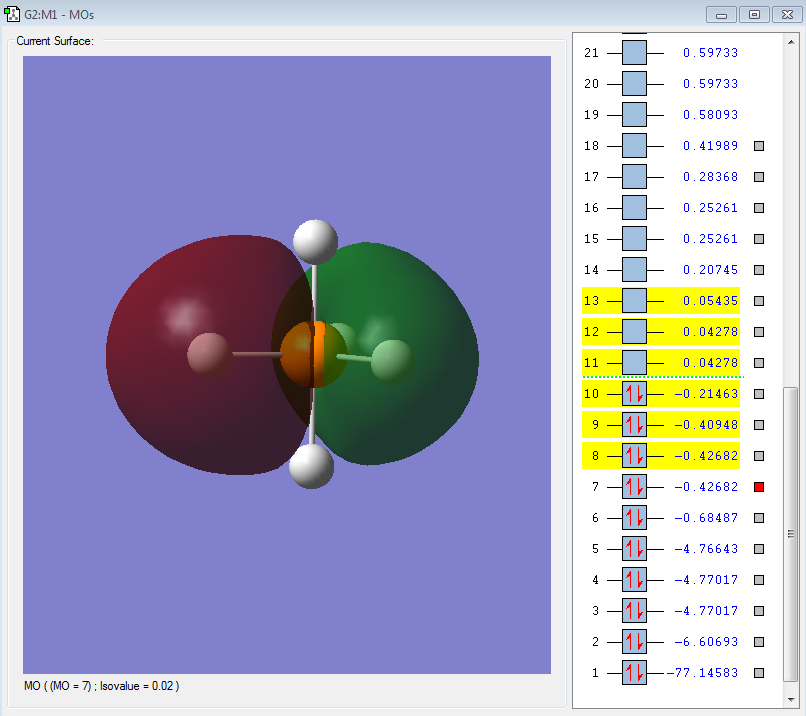
Molecular Orbitals
A PH5 molecule has 20 electrons in total, filling up 10 MOs as shown below:
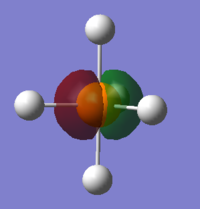
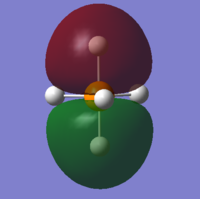
The lowest MO is so deep in energy that it cannot be seen in 3D model because it is highly penetrated inside the molecule. It may be made up by 1s orbital of P.
Also, 2s and 2p orbitals of P are too penetrated to interact with hydrogen orbitals. Below is one of the 2p orbitals.
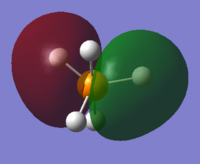
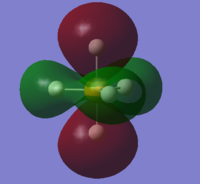
From 3s, AOs of P and H start to overlap each other to form complex MOs. Below is MO #6 made up by 3s of P and 1s of H atoms, with all AOs in phase.
3p orbitals are also mixed with hydrogen AOs but only those with orientations along the 3p lobes have contributions to MOs. #7, #8 and #9 are all bonding MOs.
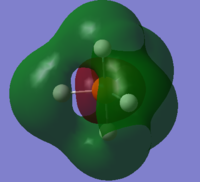
The LUMO (#10) has a very interesting shape. It is a non-bonding MO, made up by solely 5 H atoms in which the 2 axial atoms are in one phase and the other 3 equatorial atoms in opposite phase.
The HOMO (#11) and MO #12 are anti-bondings, made up by a 3p orbital and different arrangements of phases in equatorial hydrogens.
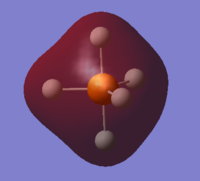
MO #13 is an anti-bonding between 3s and all of 1s orbitals in anti-phase, which forms a "cage" around the 3s!
MO #14 is the anti-bonding of 3pz and 2 axial hydrogens.
Above are all of the possible combinations between 3s3p of P and 1s of H atoms.
Higher MOs sometimes have funnier shapes!
P4 Molecule Investigation
Optimization Parameters
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -1365.44182944 a.u.
RMS Gradient Norm: 0.00014124 a.u.
Point Group: Td
Calculated Parameters
P-P bond length = 2.00 Å
P-P-P bond angle = 60°
P4 Molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000211 0.000450 YES
RMS Force 0.000125 0.000300 YES
Maximum Displacement 0.000813 0.001800 YES
RMS Displacement 0.000731 0.001200 YES
PH5 Energy calculations
E(P4)= -1365.44182944 a.u.
0.25*E(P4)= -341.36045736 a.u.
E(PH5)= -344.25491049 a.u.
E(H2)= -1.17853936 a.u.
2.5*E(H2)= -2.9463484 a.u.
ΔE=E(PH5)-[2.5*E(H2)+0.25*E(P4)]= 0.05189527 a.u.= +136.25 kJ/mol
Due to positive ΔE, the product is unstable. It is the reason why PH5 does not exist under normal conditions.