Junting1997
NH3 Molecule
Molecule name: NH3
Calculation method: RB3LYP
Basis Set: 6-31G(d.p)
final energy E(RB3LYP): -56.55776873 au
point group: C3V
N-H bond distance: 1.01798
H-N-H bond angle: 105.741
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
test molecule |
Vibration display
The optimisation is link to the file: here
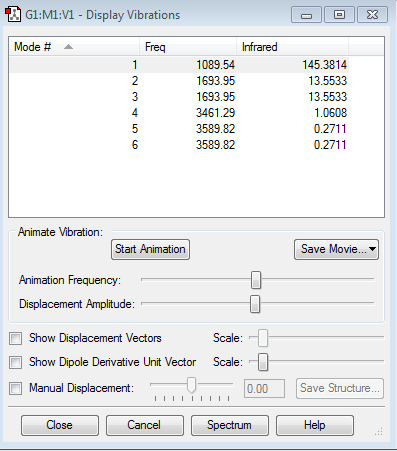
6 modes are expected from the 3N-6 rule
There are 2 different modes which are degenerated into 2 different frequencies(Frequency 1693.95 and frequency 3589.82) Mode 2 and 3 are degenerated, mode 5 and 6 are degenerated. The last 3 vibrations have very low intensity so only 2 are shown
Mode 1,2,3 are bending and Mode 4,5,6 are stretching
Mode 1 and 4 are highly symmetric
Mode 1 is knwon as the 'umbrella' mode
4 bands are expected to see in an experimental spectrum of gaseous ammonia
The charge on N-atom: -1.125 The charge on H-atoms: 0.375
I would expect the charge on N-atom to be negative, and the one on the H-atom is positive. That's because N is more electronegative than H.
N2
Molecule name: N2
Calculation method: RB3LYP
Basis Set: 6-31G(d.p)
final energy E(RB3LYP): -109.5241 au
RMS gradient: 0.00000365
point group: D*H
N-N bond distance: 1.10550 au
atom charge: 0, because they are same atoms forming a molecule
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000002 0.001800 YES RMS Displacement 0.000003 0.001200 YES Predicted change in Energy=-1.248809D-11
test molecule |
The optimisation is link to the file: here
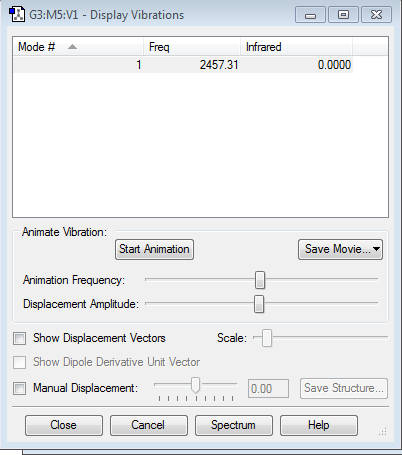
3 modes of vibration is expected. (3N-5=1 where N=2)
Display Vibration
H2
Molecule name: H2
Calculation method: RB3LYP
Basis Set: 6-31G(d.p)
final energy E(RB3LYP): -1.17859 au
RMS gradient: 0.00012170
point group: D*H
H-H bond distance: 0.74309
atom charge: 0, because the molecule is made of two same atoms.
Item Value Threshold Converged? Maximum Force 0.000211 0.000450 YES RMS Force 0.000211 0.000300 YES Maximum Displacement 0.000278 0.001800 YES RMS Displacement 0.000393 0.001200 YES
test molecule |
The optimisation is link to the file: here
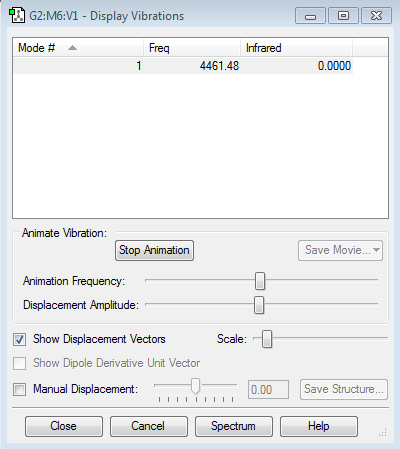
1 mode of vibration is expected. (3N-5=1 where N equals to 2)
display vibration
Haber-Bosch process
E(NH3)=-56.55776873 au
2*E(NH3)=-113.11554 au
E(N2)=-109.5241 au
E(H2)=-1.17859 au
3*E(H2)=-3.53577 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05566746 au = -146.49332 KJ/mol
ΔE is negative, so the product is more stable
Cl2
Molecule name: Cl2
Calculation method: RB3LYP
Basis Set: 6-31G(d.p)
final energy E(RB3LYP): -920.34987886 au
RMS gradient: 0.00001149
point group: D*H
Cl-Cl bond distance: 2.04155 au
atom charge: 0, because the molecule is made of two same atoms.
Item Value Threshold Converged? Maximum Force 0.000020 0.000450 YES RMS Force 0.000020 0.000300 YES Maximum Displacement 0.000056 0.001800 YES RMS Displacement 0.000079 0.001200 YES
test molecule |
1 mode of vibration is expected. (3N-5 where N=2)
The optimisation is link to the file: here
MO Theory