IRC Internuclear Distances
Introduction
This page describes a method to analyse internuclear distances during an "Intrinsic Reaction Coordinate" calculation in Gaussian. Two files are required: irc2xyz and measurements. These files must be downloaded as ".txt" files and renamed "irc2xyz.py" and "measurements.py" and kept in the same directory (folder). They can be executed cross-platform. irc2xyz converts an IRC log file to an xyz file, which measurements can use to write the internuclear distances to an Excel file. If there are any bugs or you get stuck, send me an email (tam10@ic.ac.uk) or write on the Discussion page.
News
New versions of pairdistances.py and irc2xyz have been uploaded. This should fix some incompatibility issues with Windows, but hasn't been fully tested. Tam10 (talk) 12:23, 12 February 2016 (UTC)
Pairdistances.py updated to fix a Python 3/Python 2 compatibility problem Tam10 (talk) 11:58, 18 February 2016 (UTC)
measurements added to allow measurements of angles and dihedral angles Tam10 (talk) 16:51, 18 February 2016 (UTC)
Pairdistances.py superseded by measurements.py Tam10 (talk) 11:36, 29 September 2016 (BST)
Usage
measurements will detect whether the input file is a log file or an xyz file. Any other type will fail so make sure the filename ends with either of these. In addition, only IRC log files will work. If the input is a log file, measurements will automatically call irc2xyz to convert it. This will only work, however, if irc2xyz is in the same directory as measurements and called "irc2xyz.py".
In order to use measurements, open up a Terminal (Linux/OS X) or a similar program on Windows that can execute Python script, such as cmd.exe. This can be opened by running "cmd.exe". Note that measurements cannot be opened by double clicking.
- If you are using Windows, drag python.exe from the anaconda directory in the C Drive into the terminal window (python was removed from the default build of the PCs). If you are using Linux or OS X, type "python " (spaces are important).
- Drag the measurements script file into the terminal window.
- Type " -i ", which is the input flag to tell the script to expect an input file, and drag the file to analyse into the window.
- Type " -l " (small L), which is the list flag to tell the script to expect a list of atom pairs to analyse.
The pair list must be space delimited, with each atom in a pair separated by a comma. Your line should look roughly like this:
/PATH_OF_PYTHON/python /PATH_OF_MEASUREMENTS.PY/measurements.py -i /PATH_OF_LOG_FILE/NAME_OF_LOG_FILE.log -l 1,2 2,3 3,4 4,15 15,13 13,1
Additionally, angles and dihedrals can be measured with groups of 3 or 4 atom indices respectively. For example: 1,2,3 and 1,2,3,4.
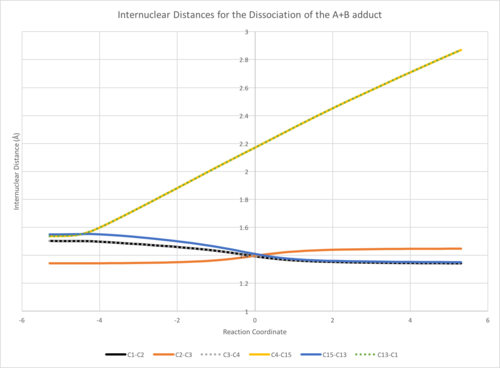
If successful you should see one or two new files, the important one being "output.csv". This can be opened with any spreadsheet app such as Excel. Inside this file there should be a table of internuclear distances with the reaction coordinate on the left.
Analysing the Output
The entire block of data can be displayed as a chart showing how the internuclear distances change over the course of the IRC.
If you're analysing a symmetric molecule, some of the lines will inevitably overlap. Either you can change the table format to make both lines visible (shown above) or rename the titles of the columns (such as C1-C2/C3-C4)