IMM2 01400724
Ammonia: NH3
Molecular Information
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -56.55776873 a.u.
RMS gradient: 0.00000485 a.u.
Point Group: C3v
N-H bond distance: 1.01798 Å
N-H bond distance literature value: 1.0124 Å [1]
H-N-H bond angle: 105.741°
H-N-H bond angle literature value: 106.670° [1]
Optimisation table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986269D-10
Optimization completed.
-- Stationary point found.
link to my NH3 file is here
NH3 |
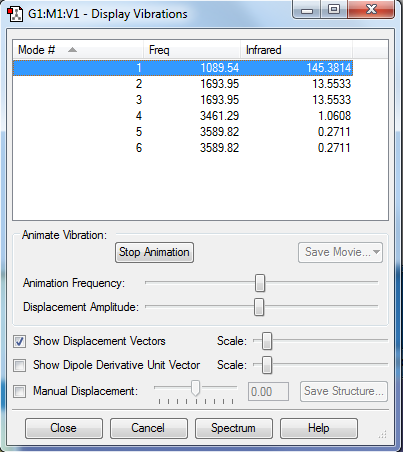
Vibrational Modes
No. of modes: 6
Degenarate: 2-3 and 5-6
Bending: 1,2,3
Stretching: 4,5,6
Highly symmetric: 4
Umbrella: 1
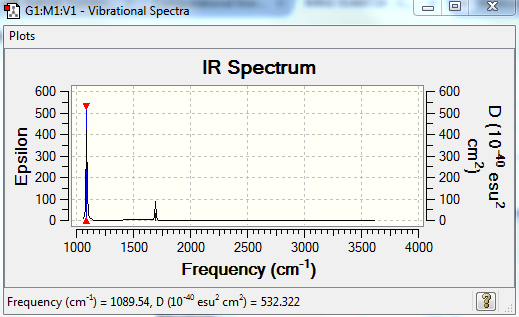
No. of bands expected: 2
- Intensities for no 4,5,6 are too small
- Number 2 and 3 are degenarate, therefore only shows one peak
IR Spectra to confirm this:
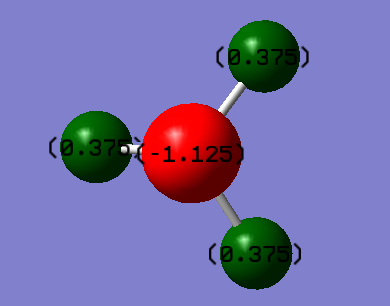
Charge Distribution
Charge on N atom: -1.125 e
Charge on H atom: 0.375 e
N has a higher electronegativity than H does, so N is able to attract the electrons more and pull them closer to its nucleus. Therefore, a negative charge was expected on the N and a positive on the H.
Diatomic Nitrogen: N2
Molecular Information
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -109.52412868 a.u.
RMS gradient: 0.00000060 a.u.
Point Group: D*H
N-N bond distance: 1.10550 Å
N-N bond distance literature value: 1.0977 Å [2]
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401111D-13
Optimization completed.
-- Stationary point found.
link to my N2 file is here
N2 |
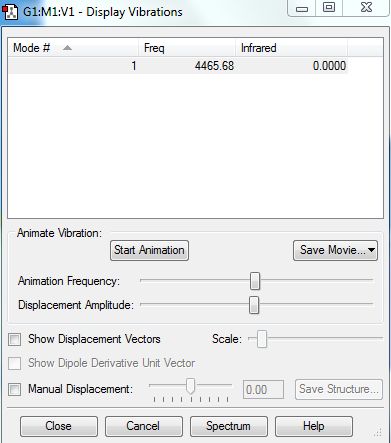
Vibrational Modes
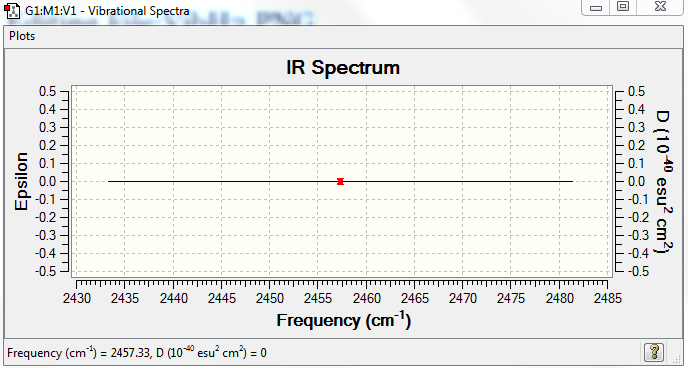
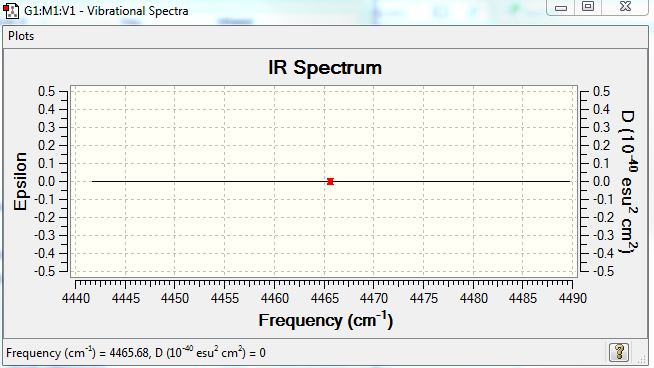
No. of bands expected: 0 (there is no dipole moment)
IR Spectra to confirm this:
Charge Distribution
No charge on N atoms, because they have the same electronegativity, therefore it has no dipole moment.
Diatomic Hydrogen: H2
Molecular Information
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -1.17853936 a.u.
RMS gradient: 0.00000017 a.u.
Point Group: D*H
H-H bond distance: 0.74279 Å
H-H bond distance literature value: 0.7414 Å [3]
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
link to my H2 file is here
H2 |
Vibrational Modes
No. of bands expected: 0 (there is no dipole moment)
IR Spectra to confirm this:
Charge Distribution
No charge on H atoms, because they have the same electronegativity, therefore it has no dipole moment.
Reaction Energies
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.1155375 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579074 a.u.
Total Energy Change: -146.4785879 kJ/mol ≈ -146.48 kJ/mol
Ammonia product is more stable than the gaseous reactants. The total energy change for the formation of ammonia is negative. Therefore energy is gained during the reaction, so it is energetically favourable to exist as ammonia.
Nitric Oxide Anion: [NO]-
Molecular Information
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -129.81016799 a.u.
RMS gradient: 0.00000011 a.u.
Point Group: C*V
N-O bond distance: 1.09197 Å
N-O bond distance literature value: 1.1508 Å [4]
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.236950D-14
Optimization completed.
-- Stationary point found.
link to my [NO]- file is here
[NO]- |
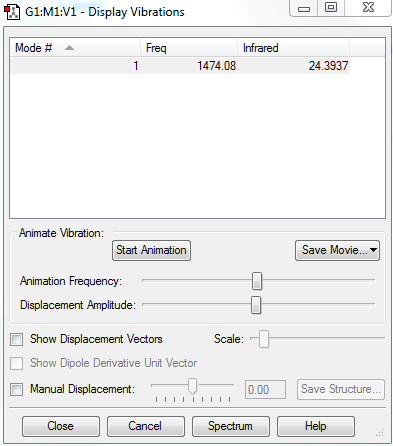
Vibrational Modes
No. of modes: 1
Stretching: 1
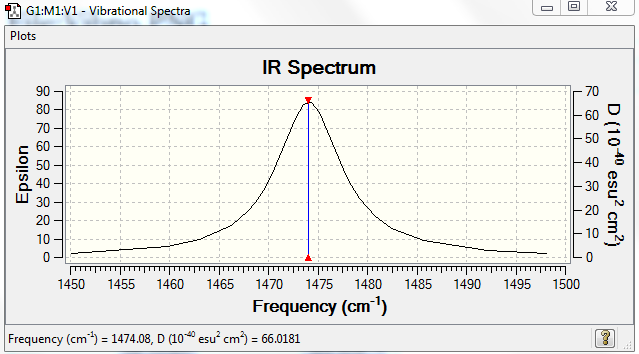
No. of bands expected: 1
IR Spectra to confirm this:
Charge Distribution
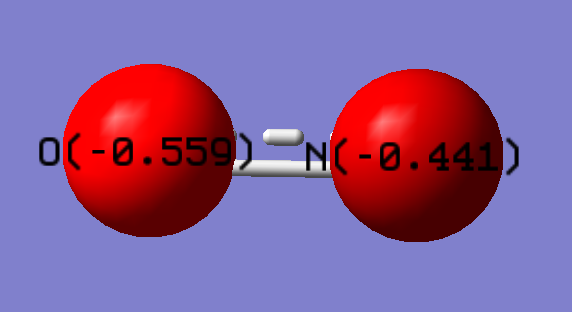
Charge on O atom: -0.559 e
Charge on N atom: -0.441 e
Negative charge was expected on both atoms. The negative charge of the whole molecule is distributed between the two atoms (delocalised). O has a more negative charge than N, because its higher electronegativity.
Molecular Orbitals
2s-2s bonding orbital: σg
- Energy: -0.72695 a.u.
- Combination of two s atomic orbitals
- Bonding
- In gerade orientation
- Occupied molecular orbital
2s-2s antibonding orbital: σu*
- Energy: -0.28482 a.u.
- Combination of two s atomic orbitals
- Antibonding
- In ungerade orientation
- Unoccupied molecular orbital
2px-2px bonding orbital: σg
- Energy: -0.05848 a.u.
- Combination of two px atomic orbitals
- Bonding
- In gerade orientation
- Occupied molecular orbital
- Green phase would be expected to be bigger in the middle, than red phases on the ends. However 2s orbitals underneath make the size of the red phases big.
2px-2px antibonding orbital: σu*
- Energy: 0.59681 a.u.
- Combination of two px atomic orbitals
- Antionding
- In ungerade orientation
- Unccupied molecular orbital
2py-2py bonding orbital: πu
- Energy: -0.05283 a.u.
- Combination of two py atomic orbitals
- Bonding
- In ungerade orientation
- HOMO: highest occupied molecular orbital
- This orbital overlap causes the high reactivity of [NO]-
2py-2py antibonding orbital: πg*
- Energy: 0.20065 a.u.
- Combination of two py atomic orbitals
- Antionding
- In gerade orientation
- LUMO: lowest unoccupied molecular orbital
Carbon Monoxide: CO
Molecular Information
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -113.30945314 a.u.
RMS gradient: 0.00001828 a.u.
Point Group: C*V
C-O bond distance: 1.13793 Å
C-O bond distance literature value: 1.1282 Å [5]
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000032 0.000450 YES
RMS Force 0.000032 0.000300 YES
Maximum Displacement 0.000012 0.001800 YES
RMS Displacement 0.000018 0.001200 YES
Predicted change in Energy=-3.956716D-10
Optimization completed.
-- Stationary point found.
link to my CO file is here
CO |
Vibrational Modes
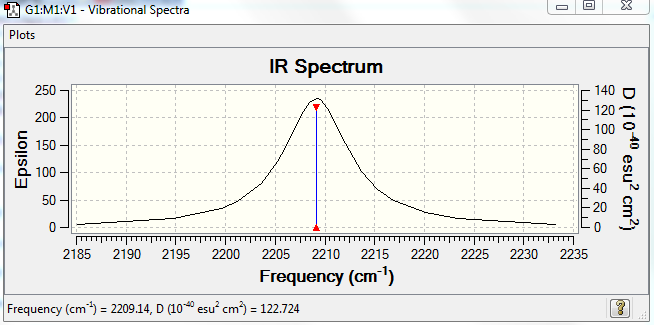
No. of modes: 1
Stretching: 1
No. of bands expected: 1
IR Spectra to confirm this:
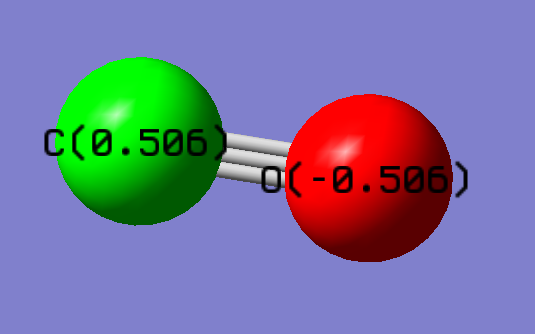
Charge Distribution
Charge on C atom: 0.506 e
Charge on O atom: -0.506 e
Negative charge was expected on O and positive on C because O has a higher electonegativity than C does. Therefore O holds the electrons closer than C.
Molecular Orbitals
2s-2s bonding orbital: σg
- Energy: -1.15791 a.u.
- Combination of two s atomic orbitals
- Bonding
- In gerade orientation
- Occupied molecular orbital
2s-2s antibonding orbital: σu*
- Energy: -0.57004 a.u.
- Combination of two s atomic orbitals
- Antibonding
- In ungerade orientation
- Unoccupied molecular orbital
2px-2px bonding orbital: σg
- Energy: -0.37145 a.u.
- Combination of two px atomic orbitals
- Bonding
- In gerade orientation
- HOMO: highest occupied molecular orbital
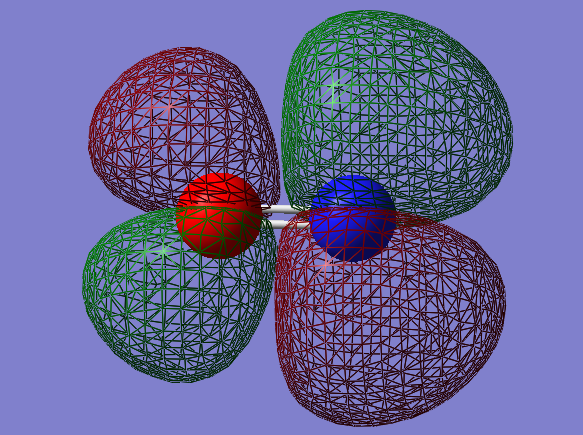
2py-2py bonding orbital: πu
- Energy: -0.46743 a.u.
- Combination of two py atomic orbitals
- Bonding
- In ungerade orientation
- Occupied
2py-2py antibonding orbital: πg*
- Energy: -0.02177 a.u.
- Combination of two py atomic orbitals
- Antionding
- In gerade orientation
- LUMO: lowest unoccupied molecular orbital
References
- ↑ 1.0 1.1 NIST Computational Chemistry Comparison and Benchmark Database NIST Standard Reference Database Number 101 Release 18, October 2016, Editor: Russell D. Johnson III https://cccbdb.nist.gov/exp2x.asp?casno=7664417
- ↑ NIST Computational Chemistry Comparison and Benchmark Database NIST Standard Reference Database Number 101 Release 18, October 2016, Editor: Russell D. Johnson III https://cccbdb.nist.gov/exp2x.asp?casno=7727379
- ↑ NIST Computational Chemistry Comparison and Benchmark Database NIST Standard Reference Database Number 101 Release 18, October 2016, Editor: Russell D. Johnson III https://cccbdb.nist.gov/exp2x.asp
- ↑ NIST Computational Chemistry Comparison and Benchmark Database NIST Standard Reference Database Number 101 Release 18, October 2016, Editor: Russell D. Johnson III https://cccbdb.nist.gov/exp2x.asp?casno=10102439
- ↑ NIST Computational Chemistry Comparison and Benchmark Database NIST Standard Reference Database Number 101 Release 18, October 2016, Editor: Russell D. Johnson III https://cccbdb.nist.gov/exp2x.asp?casno=630080&charge=0