File:Bh3 optwiki.mol
Bh3_optwiki.mol (file size: 178 bytes, MIME type: text/plain)
Warning: This file type may contain malicious code. By executing it, your system may be compromised.
3rd Year Computational Laboratory: Module 2, part 1
BH3
BH3 optimisation using 3-21G basis set
- optimised B-H bond length: 1.19 Å
- optimised H-B-H bond angle: 120.0°
BH3optimisation - summary table
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 3-21G |
| Final energy in atomic units | -26.46226338 |
| Gradient | 0.00020672 |
| Dipole moment | 0.0000 D |
| Point group | D3H |
| Duration of calculation | 24s |
BH3optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
BH3 optimisation using 6-31Gdp basis set
- optimised B-H bond length: 1.19 Å
- optimised B-H-B bond angle: 120.0°
- energy of 3-21G optimised BH3 molecule: -26.46226338 a.u.
- energy of 6-21dp optimised BH3 molecule: -26.61532264 a.u.
- difference in energy: 0.15305926 = ~402.00 kJ/mol
BH3optimisation - summary table
BH3 optimisation File Name = BH3_OPT2_631_dp File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -26.61532264 a.u. RMS Gradient Norm = 0.00021663 a.u. Imaginary Freq = Dipole Moment = 0.0000 Debye Point Group = CS Job cpu time: 0 days 0 hours 0 minutes 42.0 seconds.
BH3optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000434 0.000450 YES
RMS Force 0.000284 0.000300 YES
Maximum Displacement 0.001698 0.001800 YES
RMS Displacement 0.001114 0.001200 YES
Predicted change in Energy=-1.188111D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1914 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1914 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1914 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0001 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0001 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9999 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
BH3 frequency
BH3frequency - summary table
BH3 frequency File Name = filip_bh3_freq File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -26.61532264 a.u. RMS Gradient Norm = 0.00021659 a.u. Imaginary Freq = 0 Dipole Moment = 0.0000 Debye Point Group = C2V Job cpu time: 0 days 0 hours 0 minutes 34.0 seconds.
BH3frequency - confirmation
Item Value Threshold Converged? Maximum Force 0.000432 0.000450 YES RMS Force 0.000217 0.000300 YES Maximum Displacement 0.001697 0.001800 YES RMS Displacement 0.000848 0.001200 YES Predicted change in Energy=-1.101958D-06 Optimization completed.
Low frequencies --- -73.0837 -69.5094 -69.4394 -0.0009 -0.0007 0.0006 Low frequencies --- 1161.3810 1212.0964 1212.1646
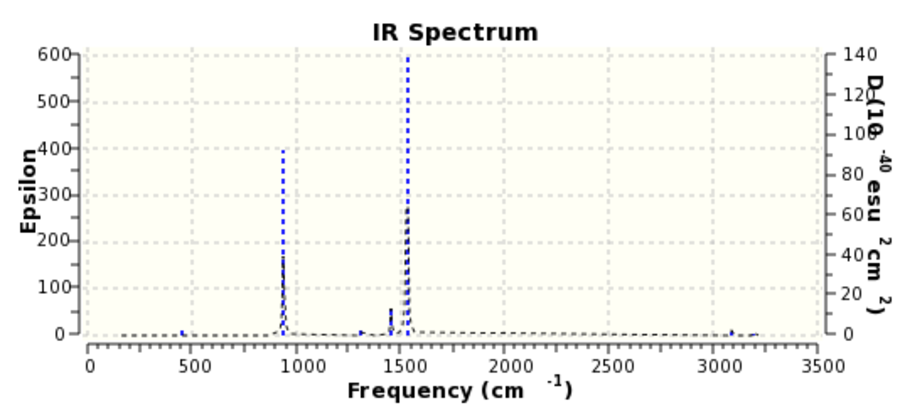
BH3 vibrational analysis
IR spectrum of BH3:
- there are only three peaks visible in the spectrum as two pairs of vibrations are degenerate so they only produce two peaks and one vibration is totally symmetric preserving the dipole. The requirement for IR active bands is that there must be a change in dipole moment and thus the interaction with electromagnetic field is possible. The last vibration gives rise to the third observed peak.
- N.B. e' are degenerate vibrations and if the symmetry was D3h, their two frequency values would be exactly the same (they vary in 3rd decimal place. Due to the C2v symmetry, they slightly vary. C2v point group could not have been avoided despite many attempts to solve the problem.
BH3 molecular orbitals
http://hdl.handle.net/10042/20492
BH3molecular orbitals - summary table
BH3 MOs File Name = BH3_MOs File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G Charge = 0 Spin = Singlet E(RB3LYP) = -7739.67280730 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 0.0002 Debye Point Group = C2V Job cpu time: 0 days 0 hours 0 minutes 27.6 seconds.
BH3molecular orbitals diagram
- the LCAO molecular orbitals look very similarly to the real ones. They lack the exact shape of the orbitals but in average, they represent the electron density quite well. If mixing was included as accurately as possible, there would be even more resemblance between them. The qualitative analysis serves as a very good approximation and proves to be useful and reliable enough to characterise smaller molecules quite quickly and precisely.
- the first core molecular orbital is not included in the diagram above due to it being too deep in energy compared to the valence orbitals
TlBr3
TlBr3optimisation
Optimised TlBr3 molecule: http://hdl.handle.net/10042/20440
- optimised Tl-Br bond length: 2.65 Å
- optimised Br-Tl-Br bond angle: 120.0°
- literature Tl-Br bond length: 2.51 Å1
[1] J. Glaser and G. Johansson, On the Structures of the Hydrated Thallium(III) Ion and its Bromide Complexes in Aqueous Solution, Acta Chemica Scandinavica A 36 (1982) 125 - 135
TlBr3optimisation - summary table
TlBr3 optimisation File Name = TlBr3optimisation_logfile File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = LANL2DZ Charge = 0 Spin = Singlet E(RB3LYP) = -91.21812851 a.u. RMS Gradient Norm = 0.00000090 a.u. Imaginary Freq = Dipole Moment = 0.0000 Debye Point Group = D3H Job cpu time: 0 days 0 hours 0 minutes 18.5 seconds.
TlBr3optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084058D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
TlBr3frequency
http://hdl.handle.net/10042/20473
TlBr3frequency - summary table
TlBr3 frequency File Name = TlBr3_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = LANL2DZ Charge = 0 Spin = Singlet E(RB3LYP) = -91.21812851 a.u. RMS Gradient Norm = 0.00000088 a.u. Imaginary Freq = 0 Dipole Moment = 0.0000 Debye Point Group = D3H Job cpu time: 0 days 0 hours 0 minutes 15.9 seconds.
TlBr3frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
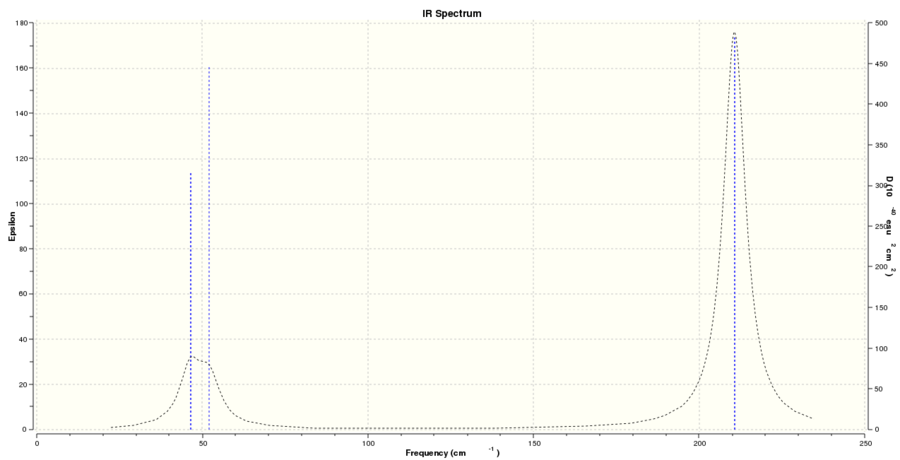
Vibrational analysis
Vibrational frequencies: 46, 46, 52, 165, 211 and 211 cm-1
BBr3
BBr3optimisation
- optimised B-Br bond length: 1.93 Å
- optimised Br-B-Br bond angle: 120.0°
Discussion
| BH3 | TlBr3 | BBr3 |
|---|---|---|
| 1.19 | 2.65 | 1.93 |
- the bond length increases in order BH3 < BBr3 < TlBr3, whereas the bond strength in the opposite direction. The bond length depends on the size of an atom, electronegativity of both elements forming the bond and whether there is a good orbital overlap. The size of the atoms present increases in order H < B < Br < Tl.
- the larger the central atom, the longer the bond. This can be seen in BBr3 (1.93 Å) and TlBr3 (2.65 Å). The same is true for the ligands, e.g. BH3 (1.19 Å) versus BBr3 (1.93 Å). This is due to the more diffuse orbitals, worse overlap and simply the atoms are further apart.
- hydrogens consist of s-type orbitals generally giving good overlap within a small volume and thus the bond lengths involving hydrogens tend to be short. Bromine is large with diffuse orbitals and tend to form weak and long bonds instead. It also has larger electronegativity pulling electrons towards itself when binding unless bound to an element with even higher electronegativity making the nature of a bond more polarised. Boron and Thallium have both the same electronegativity with Thallium being much larger and forming longer and weaker bonds. However, both bromine and thallium can extend their coordination to much higher numbers than hydrogen and boron so can form more bonds.
- a bond can be thought of as a build up of the total electon density.2 In average, the bond is the place of highest probability of finding an electon. Alternatively, it can be understood as the location of the best orbital overlap. If Gaussview does not draw bonds, it means that the corresponding structure exceeds some pre-defined length inside the program (the structure is not optimised enough). The pre-defined value is picked based on a literature value.
[2] P. Hunt, Molecular Orbitals in Inorganic Chemistry, Lecture 3, p.9
- N.B. The point group for BBr3 was determined to be C2v although the correct one is D3h. The latter could not have been achieved even with manually breaking the symmetry and trying to optimize again. The 'nosymm' keyword was not present. The same was true for further analysis of BH3 molecule (MO analysis).
BBr3optimisation - summary table
BBr3 optimisation File Name = BBR3_OPT_GEN File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = Gen Charge = 0 Spin = Singlet E(RB3LYP) = -64.43645277 a.u. RMS Gradient Norm = 0.00000384 a.u. Imaginary Freq = Dipole Moment = 0.0002 Debye Point Group = C2V Job cpu time: 0 days 0 hours 0 minutes 40.0 seconds.
BBr3optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000037 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.109348D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,4) 1.9339 -DE/DX = 0.0 !
! R2 R(2,4) 1.934 -DE/DX = 0.0 !
! R3 R(3,4) 1.934 -DE/DX = 0.0 !
! A1 A(1,4,2) 120.0021 -DE/DX = 0.0 !
! A2 A(1,4,3) 120.0021 -DE/DX = 0.0 !
! A3 A(2,4,3) 119.9957 -DE/DX = 0.0 !
! D1 D(1,4,3,2) 180.0 -DE/DX = 0.0 !
BH3 vs TlBr3
Vibrational frequencies
| BH3 | TlBr3 | Symmetry label D3h |
|---|---|---|
| 1212 | 46 | e' |
| 1212 | 46 | e' |
| 1161 | 52 | a2" |
| 2588 | 165 | a1' |
| 2722 | 211 | e' |
| 2722 | 211 | e' |
Symmetry labels
- TlBr3 analysis:
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Fre consts -- 0.1124 0.1124 0.1886
A1' E' E'
Frequencies -- 165.2685 210.6948 210.6948
Red. masses -- 78.9183 101.4032 101.4032
Frc consts -- 1.2700 2.6522 2.6522
- BH3 analysis:
A A A
Frequencies -- 1161.3810 1212.0964 1212.1646
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9958 0.9584 0.9585
A A A
Frequencies -- 2587.8129 2721.5077 2721.5171
Red. masses -- 1.0078 1.1273 1.1273
Frc consts -- 3.9765 4.9196 4.9196
Discussion
- vibrational frequency is proportional to force constant and reduced mass
- reduced mass of TlBr3 is much higher than that of BH3 and thus the vibrations are much slower compared to BH3
- the lowest vibrational frequency of TlBr3 has symmetry label e' whereas the one for BH3 has symmetry label a2". The opposite is true for the third lowest vibrational mode. The reordering occurs indeed within this first and third lowest vibrational frequency
- the two IR spectra both show three peaks and this is due to 2 pairs of vibrations being degenerate and one single vibration being totally symmetric. This leaves the spectra with 3 distinguishable IR active bands. The epsilon (molar extinction coefficient) value in the IR spectrum of BH3 is higher than in the one for TlBr3. The reason why the two spectra are similar in shape and relative distance of their peaks could lie in the fact that both molecules undergo the same types of vibrations. The difference between B and H atoms is similar to the difference between Tl and Br atom. Since both molecules have one large central atom surrounded by 3 smaller ones, the relative distance between the peaks would be comparable (in terms of the overall frequency). The difference in the actual numbers is then however reflected in the frequency we observe. As BH3 has stronger bonds and much lower reduced mass compared to TlBr3, its vibrations are at much higher frequency.
- once the optimisation is completed, all subsequent calculations and analyses involving the optimised molecule have to be performed using the same basis set and method. This basically ensures the same conditions (level of accuracy). If a different basis set or method were used for frequency analysis, the following result is quite likely to differ from the one which would be obtained if that different basis set or method were used initially to optimise the molecule.
- frequency analysis allows to calculate many useful parameters about a molecule, e.g. force constants and reduced masses leading to vibrations and more importantly the types of vibrations present. That can then be used to estimate the IR spectrum. Also, one can study the change in dipole moment and hence investigate the molecule's interactions with electromagnetic field.
- low frequencies represent the motion of the center of mass (the central atom). Since this is the heavier atom, it does not move much and so these frequencies should be low. If they are not, the molecule is probably not optimised well enough.
NH3
NH3 optimisation
NH3 optimisation: http://hdl.handle.net/10042/20501
NH3optimisation - summary table
NH3 optimisation File Name = NH3_optimisation File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -56.55776856 a.u. RMS Gradient Norm = 0.00000885 a.u. Imaginary Freq = Dipole Moment = 1.8464 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 20.9 seconds.
NH3optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629727D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
NH3 frequency
NH3 frequency: http://hdl.handle.net/10042/20502
NH3frequency - summary table
NH3 frequency File Name = NH3_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -56.55776856 a.u. RMS Gradient Norm = 0.00000878 a.u. Imaginary Freq = 0 Dipole Moment = 1.8464 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 13.6 seconds.
NH3frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000077 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.592773D-09
Optimization completed.
-- Stationary point found.
Low frequencies --- -30.8045 0.0007 0.0008 0.0008 20.2188 28.2150 Low frequencies --- 1089.5530 1694.1235 1694.1861
NH3 molecular orbitals
NH3 MOs: http://hdl.handle.net/10042/20505
NH3molecular orbitals - summary table
NH3 MOs File Name = NH3_MOs File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -56.55776852 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 1.8464 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 13.3 seconds.
NH3 charge distribution
Charge limits: Green to red = +1.0 to -1.0
Specific NBO charges for NH3
NH3BH3
NH3BH3 optimisation
NH3BH3 optimisation: http://hdl.handle.net/10042/20506
NH3BH3optimisation - summary table
NH3BH3 optimisation File Name = NH3BH3_optimisation File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -83.22469032 a.u. RMS Gradient Norm = 0.00005935 a.u. Imaginary Freq = Dipole Moment = 5.5648 Debye Point Group = C1 Job cpu time: 0 days 0 hours 1 minutes 24.4 seconds.
NH3BH3optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000121 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000508 0.001800 YES
RMS Displacement 0.000294 0.001200 YES
Predicted change in Energy=-1.611643D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0186 -DE/DX = -0.0001 !
! R2 R(2,7) 1.0186 -DE/DX = -0.0001 !
! R3 R(3,7) 1.0186 -DE/DX = -0.0001 !
! R4 R(4,8) 1.21 -DE/DX = -0.0001 !
! R5 R(5,8) 1.21 -DE/DX = -0.0001 !
! R6 R(6,8) 1.21 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.873 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.873 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0212 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.8692 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0298 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0297 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.8796 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8796 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.5972 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.8728 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.591 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.591 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.9998 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9967 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 59.9963 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0005 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.003 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 179.996 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0001 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9964 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -60.0034 -DE/DX = 0.0 !
NH3BH3 frequency
NH3BH3 frequency: http://hdl.handle.net/10042/20507
NH3BH3frequency - summary table
NH3BH3 frequency File Name = NH3BH3_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -83.22469032 a.u. RMS Gradient Norm = 0.00005931 a.u. Imaginary Freq = 0 Dipole Moment = 5.5648 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 58.7 seconds.
NH3BH3frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000113 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000583 0.001800 YES
RMS Displacement 0.000343 0.001200 YES
Predicted change in Energy=-1.712668D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- 0.0009 0.0012 0.0014 18.5248 23.7751 41.0186 Low frequencies --- 266.2845 632.2308 639.8256
NH3BH3 energy analysis
- BH3: -26.61532264 a.u.
- NH3: -56.55776856 a.u.
- BH3NH3: -83.22469032 a.u.
- ΔE=E(NH3BH3)-[E(NH3)+E(BH3)] = -0.05159912 a.u. = -135.52 kJ/mol
- the dissociation energy of the N->B dative bond is thus 135.52 kJ/mol. This is a reasonable value as single bonds are usually in a range of few hundreds kJ/mol. The actual literature value was found to be 130 kJ/mol.3
[3] S. Xie, Y. Song, and Z. Liu, Can. J. Chem. 87, 1235–1247 (2009)
3rd Year Computational Laboratory: Module 2, part 2, Ionic Liquids: Designer Solvents
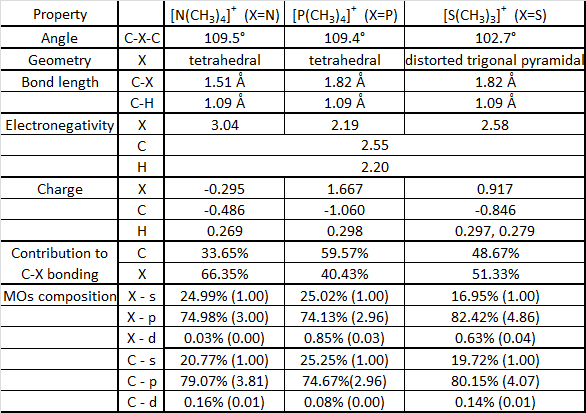
[N(CH3)4]+
[N(CH3)4]+ optimisation
[N(CH3)4]+ optimisation: http://hdl.handle.net/10042/20557
[N(CH3)4]+ optimisation - summary table
N(CH3)4 optimisation File Name = [N(CH3)4]_optimisation File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -214.18127212 a.u. RMS Gradient Norm = 0.00010472 a.u. Imaginary Freq = Dipole Moment = 2.9934 Debye Point Group = C1 Job cpu time: 0 days 0 hours 4 minutes 20.9 seconds.
[N(CH3)4]+ optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000270 0.000450 YES
RMS Force 0.000054 0.000300 YES
Maximum Displacement 0.000790 0.001800 YES
RMS Displacement 0.000209 0.001200 YES
Predicted change in Energy=-4.731250D-07
Optimization completed.
-- Stationary point found.
[N(CH3)4]+ frequency
[N(CH3)4]+ frequency: http://hdl.handle.net/10042/20584
[N(CH3)4]+ frequency - summary table
N(CH3)4 frequency File Name = [N(CH3)4]_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -214.18127212 a.u. RMS Gradient Norm = 0.00010477 a.u. Imaginary Freq = 0 Dipole Moment = 2.9934 Debye Point Group = C1 Job cpu time: 0 days 0 hours 8 minutes 52.1 seconds.
[N(CH3)4]+ frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000364 0.000450 YES
RMS Force 0.000105 0.000300 YES
Maximum Displacement 0.000825 0.001800 YES
RMS Displacement 0.000306 0.001200 YES
Predicted change in Energy=-5.687183D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -11.2532 -0.0005 -0.0001 0.0007 9.6421 13.2689 Low frequencies --- 181.9903 280.1494 287.2544
[N(CH3)4]+ molecular orbitals
[N(CH3)4]+ molecular orbitals: http://hdl.handle.net/10042/20665
[N(CH3)4]+ molecular orbitals - summary table
N(CH3)4 MOs File Name = [N(CH3)4]_MOs File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -214.18126893 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 0.0013 Debye Point Group = C1 Job cpu time: 0 days 0 hours 1 minutes 9.4 seconds.
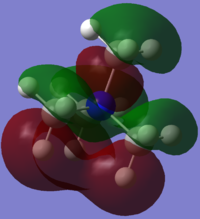
[P(CH3)4]+
[P(CH3)4]+ optimisation
[P(CH3)4]+ optimisation: http://hdl.handle.net/10042/20563
[P(CH3)4]+ optimisation - summary table
P(CH3)4 optimisation File Name = [P(CH3)4]_optimisation File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -500.82700332 a.u. RMS Gradient Norm = 0.00003441 a.u. Imaginary Freq = Dipole Moment = 3.7357 Debye Point Group = C1 Job cpu time: 0 days 0 hours 4 minutes 27.7 seconds.
[P(CH3)4]+ optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000075 0.000450 YES
RMS Force 0.000026 0.000300 YES
Maximum Displacement 0.001055 0.001800 YES
RMS Displacement 0.000294 0.001200 YES
Predicted change in Energy=-9.220745D-08
Optimization completed.
-- Stationary point found.
[P(CH3)4]+ frequency
[P(CH3)4]+ frequency: http://hdl.handle.net/10042/20591
[P(CH3)4]+ frequency - summary table
P(CH3)4 frequency File Name = [P(CH3)4]_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -500.82700332 a.u. RMS Gradient Norm = 0.00003444 a.u. Imaginary Freq = 0 Dipole Moment = 3.7357 Debye Point Group = C1 Job cpu time: 0 days 0 hours 8 minutes 20.7 seconds.
[P(CH3)4]+ frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000083 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.002272 0.001800 NO
RMS Displacement 0.000935 0.001200 YES
Predicted change in Energy=-1.113948D-07
- N.B. for frequency analysis, only the forces are required to converge. Moreover, the difference between the actual value and the threshold value is not that large.
Low frequencies --- -17.5556 0.0018 0.0024 0.0031 2.2658 15.3518 Low frequencies --- 155.1983 184.5950 192.4405
[P(CH3)4]+ molecular orbitals
[P(CH3)4]+ molecular orbitals: http://hdl.handle.net/10042/20678
[P(CH3)4]+ molecular orbitals - summary table
P(CH3)4 MOs File Name = [P(CH3)4]_MOs File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -500.82699801 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 0.0021 Debye Point Group = C1 Job cpu time: 0 days 0 hours 1 minutes 11.7 seconds.
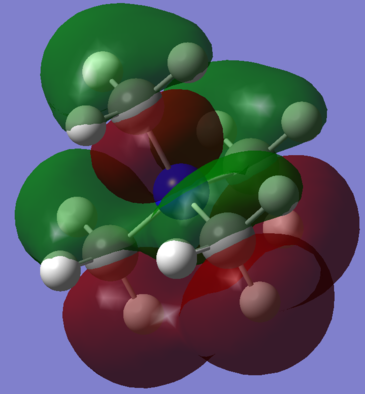
[S(CH3)3]+
[S(CH3)3]+ optimisation
[S(CH3)3]+ optimisation: http://hdl.handle.net/10042/20564
[S(CH3)3]+ optimisation - summary table
S(CH3)3 optimisation File Name = [S(CH3)3]_optimisation File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -517.68327867 a.u. RMS Gradient Norm = 0.00006722 a.u. Imaginary Freq = Dipole Moment = 3.1533 Debye Point Group = C1 Job cpu time: 0 days 0 hours 2 minutes 54.2 seconds.
[S(CH3)3]+ optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000145 0.000450 YES
RMS Force 0.000039 0.000300 YES
Maximum Displacement 0.001734 0.001800 YES
RMS Displacement 0.000500 0.001200 YES
Predicted change in Energy=-2.931400D-07
Optimization completed.
-- Stationary point found.
[S(CH3)3]+ frequency
[S(CH3)3]+ frequency: http://hdl.handle.net/10042/20605
[S(CH3)3]+ frequency - summary table
S(CH3)3 frequency File Name = [S(CH3)3]_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -517.68327867 a.u. RMS Gradient Norm = 0.00006719 a.u. Imaginary Freq = 0 Dipole Moment = 3.1533 Debye Point Group = C1 Job cpu time: 0 days 0 hours 3 minutes 59.0 seconds.
[S(CH3)3]+ frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000270 0.000450 YES
RMS Force 0.000067 0.000300 YES
Maximum Displacement 0.001798 0.001800 YES
RMS Displacement 0.000647 0.001200 YES
Predicted change in Energy=-3.305774D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -13.7961 -11.8440 0.0012 0.0039 0.0040 22.1581 Low frequencies --- 159.2984 195.2522 199.2714
[S(CH3)3]+ MOs
[S(CH3)3]+ MOs: http://hdl.handle.net/10042/20694
[S(CH3)3]+ MOs - summary table
S(CH3)3 MOs File Name = [S(CH3)3]_MOs File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -517.68327822 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 0.9640 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 42.2 seconds.
Discussion
General analysis
- Both N and P atoms adopt tetrahedral geometry to minimize the steric crowding of methyl groups which are thus in the same environment. This is confirmed by inspection of C-X-C bond angles (109.5° and 109.4° respectively compared to 109.7° for tetrahedral arrangement). On the other hand, S is found to be in a trigonal pyramidal structure due to its lone pair which is quite large and pushes the methyl groups closer together by a lot reducing the C-S-C from normal trigonal pyramidal (~107°) to slightly distorted molecular structure with bond angle 102.7°.
- The shortest and strongest bond among the three C-X bonds is between nitrogen and carbon. P and S atoms form weaker bonds mainly due to them being larger atoms with diffuse orbitals resulting in smaller orbital overlap. Only N is in the same period as C. The literature values for C-N (ammonium salt)4, C-P5 and C-S5 are 1.50 Å, 1.87 Å and 1.81 Å respectively. The computed values fit this quite well.
- The charge distribution in these three systems can be rationalized based on Pauling electronegativity which increases in order P < H < C < S < N and effective nuclear charge of every atom. The larger atomic number, the higher Zeff and the more electron density would be attracted to this atom. The formal oxidation state of all hydrogens is (+I), the one of all carbons is (-IV), nitrogen and phosphorus carry (+III) and sulphur (+II) oxidation number respectively. Although each complex has +1 charge, in every structure it is distributed differently among the atoms so that the minimum energy is found and the charge is stabilized best.
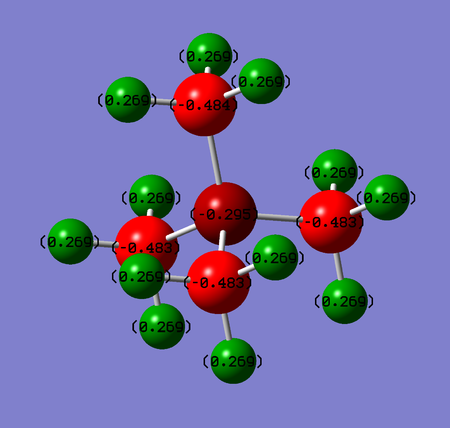
- In [N(CH3)4]+, the nitrogen atom is so electronegative that it inductively pulls electron density away from carbons. It does so to such extent that the charge on nitrogen lowers to -0.295 and all carbons therefore experience much lower negative charge (-0.483).
- In [P(CH3)4]+, the phosphorus atom is now less electronegative than carbon and hence carries high positive charge of +1.667. On the contrary, the charge of each carbon decreased dramatically to -1.060 because of incoming electron density from phosphorus.
- In [S(CH3)3]+, the electronegativities of sulphur and carbon are comparable. The positive charge in the complex would be mostly stabilized on sulphur but the actual charge is not +1 (but +0.917) as sulphur itself withdraws little electron density from carbons due to its marginally higher electronegativity. The carbon electon density is somewhat lowered by this effect displaying lower negative charge (-0.846) than in the complex with electopositive phosphorus but higher than in the complex with electronegative nitrogen.
- The charge for hydrogens does not change by much across these three complexes oscillating in a range of +0.269 to +0.298.
- The charge distribution completely relates to the contribution of each of the two atoms to the C-X bond. The atom with higher electronegativity has got higher electron density around itself and thus contributes to the bond more than its bonding partner. The higher the difference in electronegativity, the bigger the difference in contributions:
- [N(CH3)4]+ ΔχC-N=0.49, contribution factor ~34:66
- [P(CH3)4]+ ΔχC-P=0.36, contribution factor ~60:40
- [S(CH3)3]+ ΔχC-S=0.03, contribution factor ~49:51
- The p-type orbitals dominate the MO character of all carbons and heteroatoms whereas MOs of hydrogens are almost exclusively formed from s-type orbitals. The highest contribution of p-type character is observed on sulphur atom (s:p = ~1:5). Apart from p-type, s-type orbitals also participate in carbons and heteroatoms but to a much lesser extent (s:p = ~1:3 - ~1:4). Only traces of d-type orbitals have been indicated.
- Although the positive charge is often assigned to the nitrogen atom in [N(CH3)4]+, according to the charge distribution analysis, this is not a very realistic situation. Nitrogen is too electronegative and pulls electron density towards itself. A positive charge spread mainly on hydrogens and somewhat less on carbons would be a better representation for this complex. It is always better to designate the positive charge to the whole molecule rather than a separate single atom unless charge distribution analysis has already been done and verified. In addition, R groups (R=alkyl) are electron releasing via induction always decreasing the partial charge on central nitrogen. Consequently, depicting a positive charge on this atom is imprecise.
[4] F. H. Allen, et. al. Tables of bond Lengths determined by X-Ray and Neutron Diffraction. Part 1. Bond Lengths in Organic Compounds, J. Chem. Soc. II 1987, S1-S19.
[5] R. C. Weast, Handbook of Chemistry & Physics (65th ed.), CRC Press, ISBN 0-8493-0465-2.
Interactions in molecular orbitals
[N(CH3)3(CH2OH)]+
[N(CH3)3(CH2OH)]+ optimisation
[N(CH3)3(CH2OH)]+ optimisation: http://hdl.handle.net/10042/20716
[N(CH3)3(CH2OH)]+ optimisation - summary table
OH optX File Name = OH opt2 File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -289.39472261 a.u. RMS Gradient Norm = 0.00001557 a.u. Imaginary Freq = Dipole Moment = 1.8145 Debye Point Group = C1 Job cpu time: 0 days 0 hours 19 minutes 23.1 seconds.
[N(CH3)3(CH2OH)]+ optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000030 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.000924 0.001800 YES
RMS Displacement 0.000263 0.001200 YES
Predicted change in Energy=-2.595792D-08
Optimization completed.
-- Stationary point found.
[N(CH3)3(CH2OH)]+ frequency
[N(CH3)3(CH2OH)]+ frequency: http://hdl.handle.net/10042/20720
[N(CH3)3(CH2OH)]+ frequency - summary table
OH freqX File Name = OH freq2 File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -289.39472261 a.u. RMS Gradient Norm = 0.00001549 a.u. Imaginary Freq = 0 Dipole Moment = 1.8145 Debye Point Group = C1 Job cpu time: 0 days 0 hours 12 minutes 5.7 seconds.
[N(CH3)3(CH2OH)]+ frequency - confirmation
Item Value Threshold Converged?
Maximum Force 0.000076 0.000450 YES
RMS Force 0.000015 0.000300 YES
Maximum Displacement 0.000779 0.001800 YES
RMS Displacement 0.000282 0.001200 YES
Predicted change in Energy=-2.811110D-08
Optimization completed.
-- Stationary point found.
Low frequencies --- -8.3571 0.0009 0.0010 0.0013 9.3119 23.2991 Low frequencies --- 135.6194 212.4618 256.7404
[N(CH3)3(CH2OH)]+ molecular orbitals
[N(CH3)3(CH2OH)]+ molecular orbitals: http://hdl.handle.net/10042/20726
[N(CH3)3(CH2OH)]+ molecular orbitals - summary table
N(CH3)3(CH2OH)+ MOs File Name = OH MOs2 File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -289.39471439 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 2.1373 Debye Point Group = C1 Job cpu time: 0 days 0 hours 1 minutes 47.0 seconds.
[N(CH3)3(CH2CN)]+
[N(CH3)3(CH2CN)]+ optimisation
[N(CH3)3(CH2CN)]+ optimisation: http://hdl.handle.net/10042/20569
[N(CH3)3(CH2CN)]+ optimisation - summary table
[N(CH3)3(CH2CN)]+ optimisation File Name = [N(CH3)3(CH2CN)]_optimisation File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -306.39377650 a.u. RMS Gradient Norm = 0.00001756 a.u. Imaginary Freq = Dipole Moment = 13.0927 Debye Point Group = C1 Job cpu time: 0 days 0 hours 18 minutes 17.8 seconds.
[N(CH3)3(CH2CN)]+ optimisation - confirmation
Item Value Threshold Converged?
Maximum Force 0.000044 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000561 0.001800 YES
RMS Displacement 0.000155 0.001200 YES
Predicted change in Energy=-1.656738D-08
Optimization completed.
-- Stationary point found.
[N(CH3)3(CH2CN)]+ frequency
[N(CH3)3(CH2CN)]+ frequency: http://hdl.handle.net/10042/20643
[N(CH3)3(CH2CN)]+ frequency - summary table
[N(CH3)3(CH2CN)]+ frequency File Name = [N(CH3)3(CH2CN)]_frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -306.39377650 a.u. RMS Gradient Norm = 0.00001761 a.u. Imaginary Freq = 0 Dipole Moment = 13.0926 Debye Point Group = C1 Job cpu time: 0 days 0 hours 12 minutes 7.2 seconds.
[N(CH3)3(CH2CN)]+ frequency - confirmation
Item Value Threshold Converged? Maximum Force 0.000056 0.000450 YES RMS Force 0.000018 0.000300 YES Maximum Displacement 0.001834 0.001800 NO RMS Displacement 0.000605 0.001200 YES Predicted change in Energy=-3.139986D-09
Low frequencies --- -0.0002 0.0007 0.0009 5.4931 11.0165 16.8454 Low frequencies --- 93.2738 154.2316 213.7761
[N(CH3)3(CH2CN)]+ molecular orbitals
[N(CH3)3(CH2CN)]+ molecular orbitals: http://hdl.handle.net/10042/20710
[N(CH3)3(CH2CN)]+ molecular orbitals - summary table
[N(CH3)3(CH2CN)]+ MOs File Name = [N(CH3)3(CH2CN)]_MOs File Type = .log Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -306.39376999 a.u. RMS Gradient Norm = a.u. Imaginary Freq = Dipole Moment = 5.7640 Debye Point Group = C1 Job cpu time: 0 days 0 hours 1 minutes 43.6 seconds.
Discussion
Effects of different substituents on charge distribution
 |
 |
|
|---|---|---|
| [N(CH3)4]+ | [N(CH3)3CH2OH]+ | [N(CH3)3CH2CN]+ |
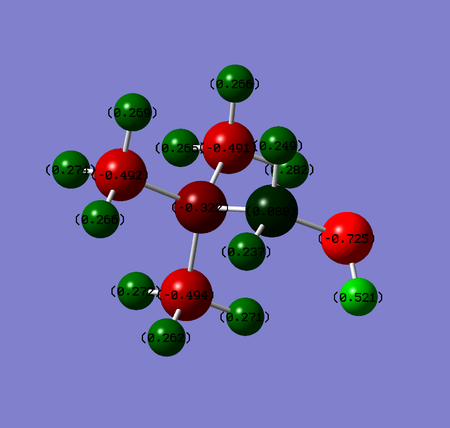
- The picture in the middle confirmes that OH group is electron withdrawing by induction and electron donating via resonance due to oxygen lone pairs. The resonance effect generally predominates and this can be seen in this case as well. The electron density on nitrogen, three carbons and hydrogens (especially the closest ones to OH group) increased dramatically after introducing the OH substituent. The only element experiencing a stronger inductive than resonance effect of oxygen is carbon adjacent to OH group. The electronegative oxygen pulls electron density away from it and increases its charge from -0.483 to +0.088.
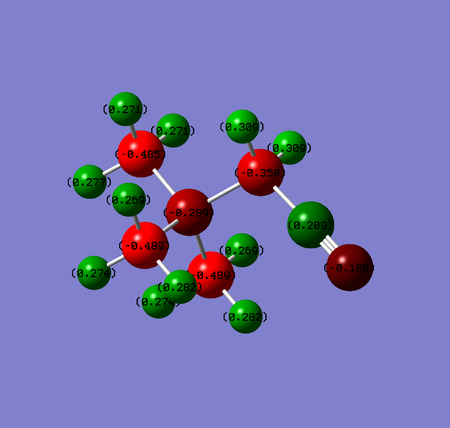
- CN group however is an electron withdrawing group by resonance as the nitrogen atom tries to break a bond with carbon in order to gain a lone pair. This is a result of high electronegativity of nitrogen. The effect can be justified by comparing charge distributions with and without CN substituent. In case a substituent is present, all atoms except three carbons further away from cyano group (and these decreased only slightly; by 0.004-0.006) lost some electron density and their charges have increased.
HOMO-LUMO comparison
- The shape of the HOMO-LUMO orbitals involving substituents changes dramatically as the HOMO contains only half of the atomic orbitals whereas LUMO consists of all of them. LUMO is thus more delocalised. Also, the number of nodal planes and atomic nodes increases making the character of the molecular orbital more antibonding and less likely to be occupied. For [N(CH3)4]+, the number of atomic orbitals forming the molecular orbital is significant in both cases but the character changes markedly. 5 nodal planes in LUMO (some of them spherical) compared to 3 in HOMO make the LUMO higher in energy.
- The HOMO-LUMO energy gaps have been calculated to be 0.44609, 0.34639 and 0.31867 a.u. for [N(CH3)4]+, [N(CH3)3CH2OH]+ and [N(CH3)3CH2CN]+ respectively. It is the highest difference between two adjacent energy levels in the whole molecular orbitals diagram except one gap which is slightly larger but occurs towards the top of the diagram and is chemically unimportant(above level 130). As both HOMO and LUMO are antibonding, the more elctron density present, the higher the orbital will be in energy due to stronger antibonding interactions. This correlates with the data as the electron releasing hydroxyl group is highest in energy followed by unsubstituted [N(CH3)4]+ and electron withdrawing cyano group. The reason why the latter two are swapped in HOMOs may simply lie in the fact that the HOMO of [N(CH3)4]+ is 21st level whereas the one of N(CH3)3CH2CN]+ is 27th.
- The larger the gap present, the harder it is to promote an electron from HOMO to LUMO and the less reactive and more stable the compound is. The magnitude of the energy gap also influences magnetic properties and colour not applicable to these complexes though. The smaller gap for substituted complexes means they are more likely to react with incoming electrophiles and nucleophiles (depending on charge at particular atom which has been discussed earlier). According to the data, cyano group is slightly more reactive than hydroxyl group. For the overall reactivity, however, the actual energy also has to be taken into account as a molecule with high HOMO-LUMO energy levels is unlikely to undergo addition reactions whereas the one with low lying HOMO-LUMO orbitals will be less prone to elimination reactions.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Dimensions | User | Comment | |
|---|---|---|---|---|
| current | 12:57, 8 October 2012 | (178 bytes) | Fs1310 (talk | contribs) |
You cannot overwrite this file.
File usage
There are no pages that use this file.