Ferdiemodeling2:Modeling2
introduction to molecular modeling
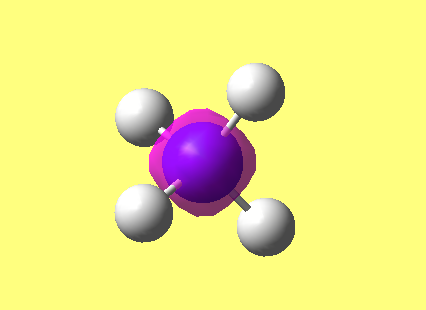
NH3
image of NH |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -56.55776873 a.u. |
| RMS Gradient Norm | 0.00000485 a.u. |
| Point Group | C3V |
| N-H bond length | 1.01798Å |
| H-N-H Angel | 105.741° |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000004 | 0.000450 | YES |
| RMS | Force | 0.000004 | 0.000300 | YES |
| Maximum | Displacement | 0.000072 | 0.001800 | YES |
| RMS | Displacement | 0.000035 | 0.001200 | YES |
| mode | Frequency | Infrared | IR active | type |
|---|---|---|---|---|
| 1 | 1089.54 | 145.3814 | yes | bending |
| 2 | 1693.95 | 13.5533 | yes | bending |
| 3 | 1693.95 | 13.5533 | yes | bending |
| 4 | 3461.29 | 1.0608 | yes | stretching |
| 5 | 3589.82 | 0.2711 | yes | stretching |
| 6 | 3589.82 | 0.2711 | yes | stretching |
- 6 modes are expected when using the 3N-6 rule
- modes 2 and 3 are degenerate and modes 5 and 6 are degenerate
- there will be 2 peaks in the absorbance spectrum as although all modes are IR active modes 4,5,6 have very low intensities which are proportional to the intensity in the spectrum and as they are so small they will be drowned out by noise in an experimental determined spectra as as modes 2 and 3 are degenerate they will occur at the same frequency
- modes 1, 2 ,and 3 are bending vibrations
- modes 4, 5, and 6 are bond stretching vibrations
- mode 1 is known as the umbrella mode
- mode 4 is the highly symmetric mode
- the N atom is predicted to have a charge of -1.125 and the H atoms have a charge of 0.375 which means that the molecule has an overall charge of 0
- we would expect that N has a negative change and so H has positive charge as it is more N is more electronegative than H and that the overall charge to be 0
data file
N2
image of N |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -109.52412868 a.u. |
| RMS Gradient Norm | 0.00000060a.u. |
| Point Group | D*H |
| N-N bond length | 1.10550Å |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000001 | 0.000450 | YES |
| RMS | Force | 0.000001 | 0.000300 | YES |
| Maximum | Displacement | 0.000000 | 0.001800 | YES |
| RMS | Displacement | 0.000000 | 0.001200 | YES |
| mode | Frequency | Infrared | IR active |
|---|---|---|---|
| 1 | 2457.33 | 0.0000 | no |
there is no IR activity as it is a linear molecule
data file
molecular orbitals:
MO1
1s σ bonding orbitals E=-14.44676a.u.
MO2
1s σ* anti-bonding orbitals E=-14.44512a.u.
MO3
2s σ bonding orbital E=-1.12383a.u.
MO4
2s σ* anti-bonding orbital E=-0.55342a.u.
MO5
2p π bonding orbital degenerate with MO6 E=-0.462540a.u.
MO6
2p π bonding orbital degenerate with MO5 E=-0.462540a.u.
MO7
2p σ bonding orbital (HOMO) E=-0.42688a.u. (non degenerate as along the axis of the bond) this dose not exactly have the same shape as expected due to there being some s character due to mixing
MO8
2p π* anti-bonding orbital (LUMO) degenerate with MO9 E=-0.02412a.u. this dose not exactly have the same shape as expected due to there being some d characteristic in the p orbitals due to the polarization function chosen
MO9
2p π* anti-bonding orbital (LUMO) degenerate with MO8 E=-0.02412a.u. this dose not exactly have the same shape as expected due to there being some S character due to mixing
MO10
2p σ* anti-bonding bonding orbital E=0.41366a.u. (non degenerate as along the axis of the bond) this dose not exactly have the same shape as expected due to there being some S character due to mixing however not as influential as in MO7 due to there being being destructive interference between the molecules
data file
H2
image of H |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -1.17853936 a.u. |
| RMS Gradient Norm | 0.00000017a.u. |
| Point Group | D*H |
| H-H bond length | 0.74279Å |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000000 | 0.000450 | YES |
| RMS | Force | 0.000000 | 0.000300 | YES |
| Maximum | Displacement | 0.000000 | 0.001800 | YES |
| RMS | Displacement | 0.000001 | 0.001200 | YES |
| mode | Frequency | Infrared | IR active |
|---|---|---|---|
| 1 | 4465.68 | 0.0000 | no |
this molecule is non IR active as it is linear and so has no dipole change
data file
Harbor-Bosh reaction
2N2 (g) + 3H2 (g) → 2NH3 (g)
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.11553746 a.u.
E(N2)=-109.52412868 a.u.
E(H2)=-1.17853936 a.u.
3*E(H2)=-3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557907 a.u.
ΔE = -146.47848285 kJ/mol
therefore ΔE = -146.48 kJ/mol
the NH3 is more stable than the gaseous reactants as ΔE is negative meaning that the reaction is exothermic, however there is an decrease in entropy meaning that this reaction will only occur below a certain temperature. As can be seen from the literature of -46210 J/mole [1] the value that is calculated would be expected to be an exothermic.
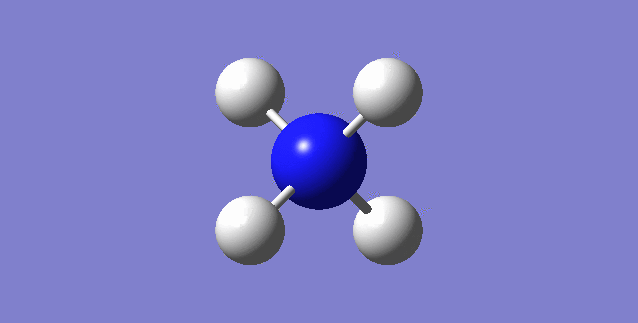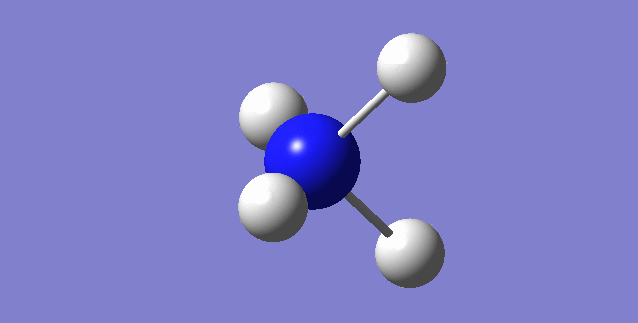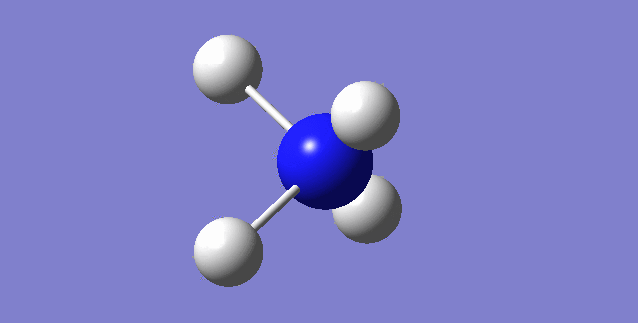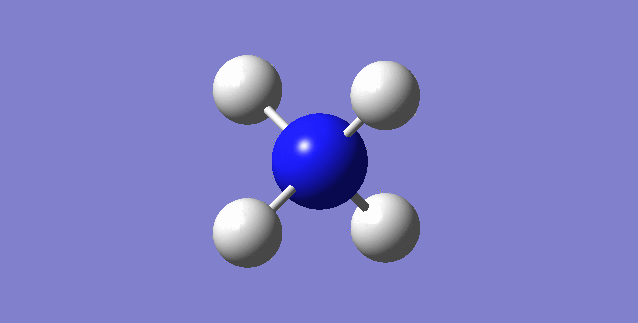
[NH4]+
image of [NH4]+
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -56.90586436 a.u. |
| RMS Gradient Norm | 0.00011991 a.u. |
| Point Group | TD |
| N-H bond length | 1.02761Å |
| H-N-H Angle | 109.47122° |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000232 | 0.000450 | YES |
| RMS | Force | 0.000124 | 0.000300 | YES |
| Maximum | Displacement | 0.000537 | 0.001800 | YES |
| RMS | Displacement | 0.000287 | 0.001200 | YES |
| mode | Frequency | Infrared | IR active | type |
|---|---|---|---|---|
| 1 | 1496.46 | 181.0390 | yes | bending |
| 2 | 1496.46 | 181.0390 | yes | bending |
| 3 | 1496.46 | 181.0390 | yes | bending |
| 4 | 1726.51 | 0.0000 | no | bending |
| 5 | 1726.51 | 0.0000 | no | bending |
| 6 | 3366.56 | 0.0000 | no | stretching |
| 7 | 3491.44 | 197.7368 | yes | stretching |
| 8 | 3491.44 | 197.7368 | yes | stretching |
| 9 | 3491.44 | 197.7368 | yes | stretching |
charges on [NH4]+ atoms
| atoms | charges |
|---|---|
| N | -0.999 |
| H | 0.500 |
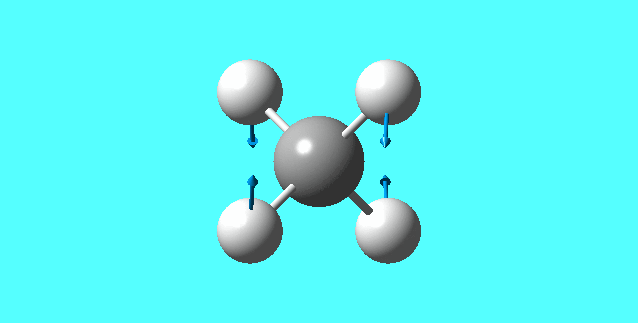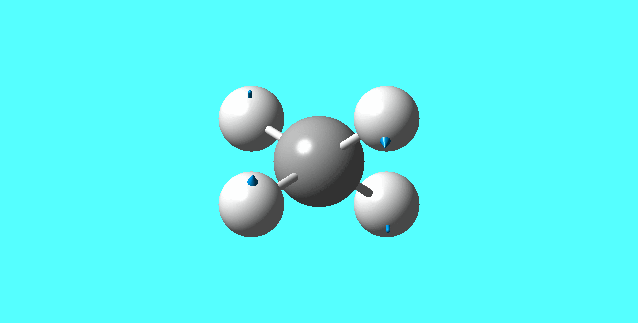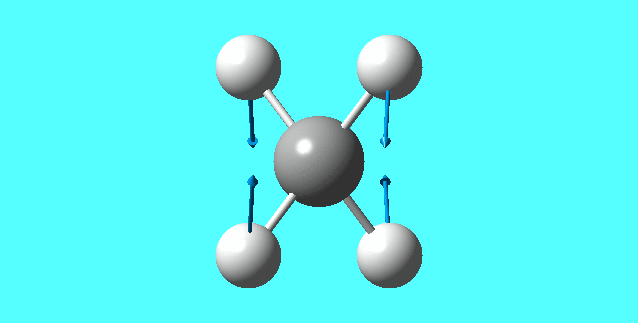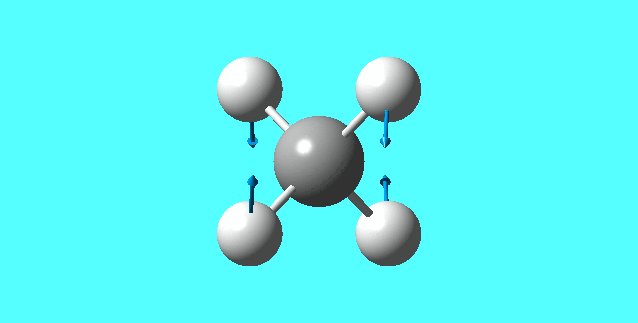
modes of vibration for [NH4]+:
mode 1
mode 2
mode 3
mode 4
mode 5
mode 6
mode 7
mode 8
mode 9
Molecular orbitals
Molecular orbital energies
| MO | occupancy | Energy (a.u.) |
|---|---|---|
| 15 | 0 | 0.5405978170 |
| 14 | 0 | 0.5405978170 |
| 13 | 0 | 0.5405978170 |
| 12 | 0 | 0.3325906440 |
| 11 | 0 | 0.3325906440 |
| 10 | 0 | 0.3325906440 |
| 9 | 0 | -0.1284344420 |
| 8 | 0 | -0.1284344420 |
| 7 | 0 | -0.1284344420 |
| 6 | 0 | -0.2100277800 |
| 5 | 2 | -0.8247061880 |
| 4 | 2 | -0.8247061880 |
| 3 | 2 | -0.8247061880 |
| 2 | 2 | -1.2480757300 |
| 1 | 2 | -14.7152516000 |
molecular orbital diagrams
MO1
in the first molecular orbital only the 1s orbital in the N is involved this is an anti bonding orbital
MO2
the nitrogen atom contributes an a 2s atomic orbital and the hydrogen atom contributes a 1s atomic orbital causing the formation of a σ bonding orbital
MO3-5
the nitrogen atom contributes an a 2p atomic orbital and the hydrogen atom contributes a 1s atomic orbital causing the formation of degenerate π bonding orbitals witch are the HOMO
MO6
the nitrogen atom contributes an a 2s atomic orbital and the hydrogen atom contributes a 1s atomic orbital causing the formation of a σ* anti-bonding orbital which is the LUMO
MO7-9
the nitrogen atom contributes an a 2p atomic orbital and the hydrogen atom contributes a 1s atomic orbital causing the formation of degenerate π* anti-bonding orbitals
MO10 and above

data file
File:FERDIE KRAMMER -NH4-+ 1 2.LOG
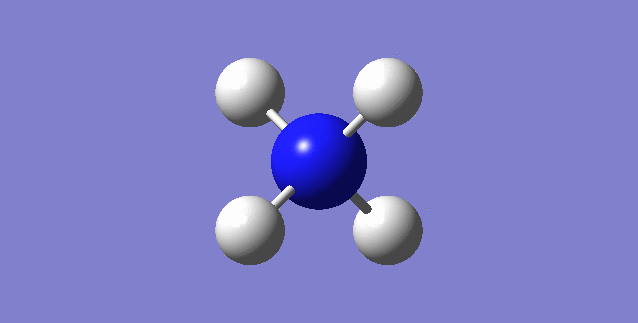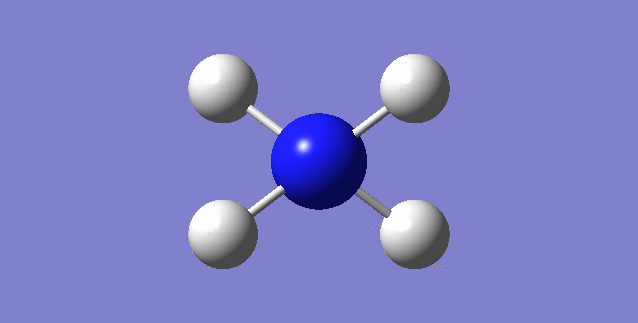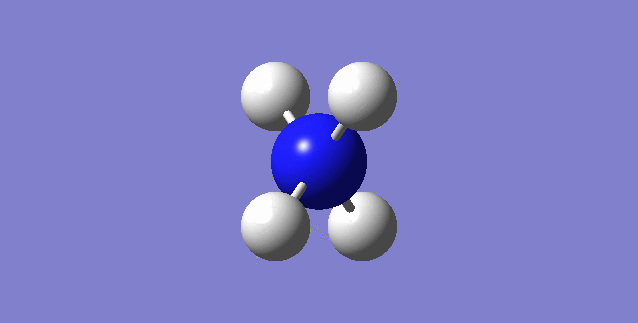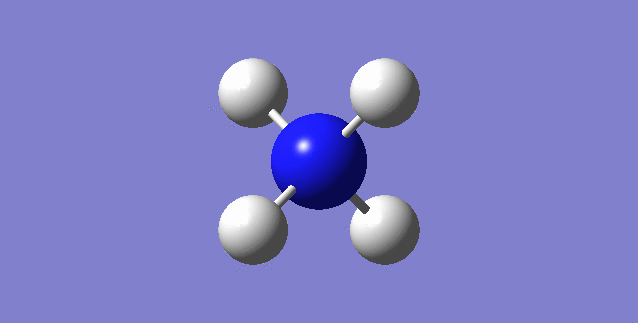
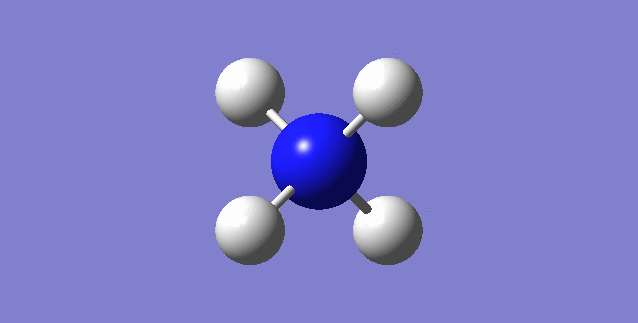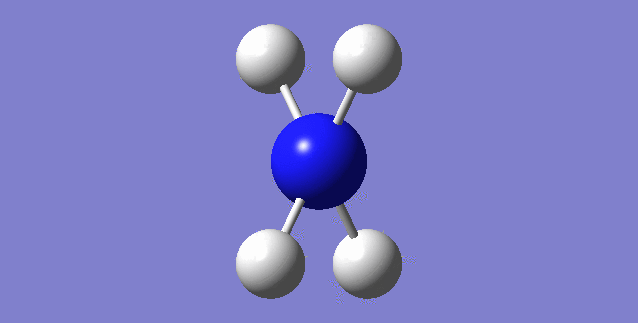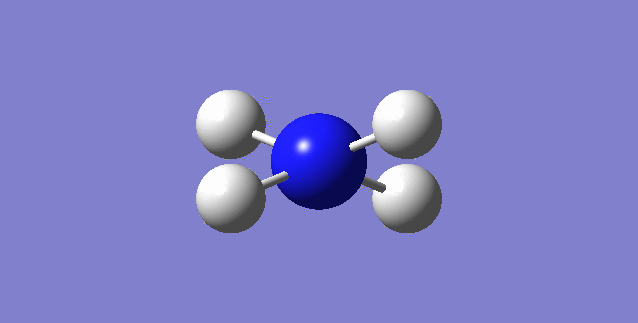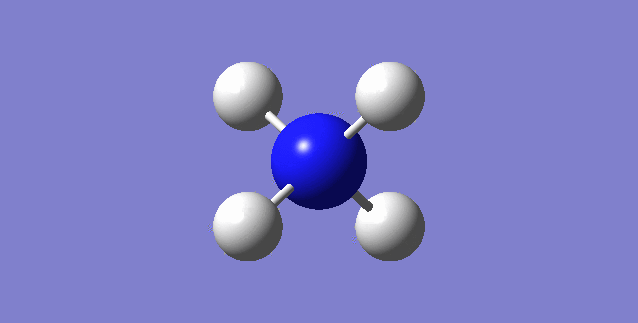
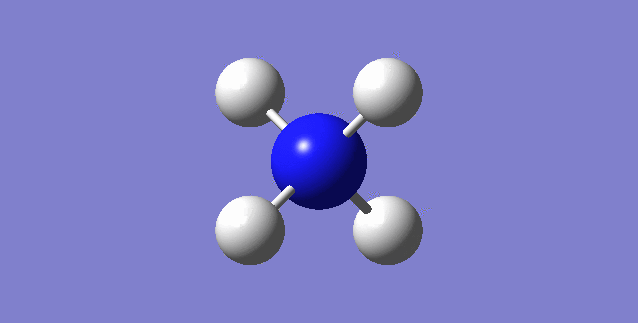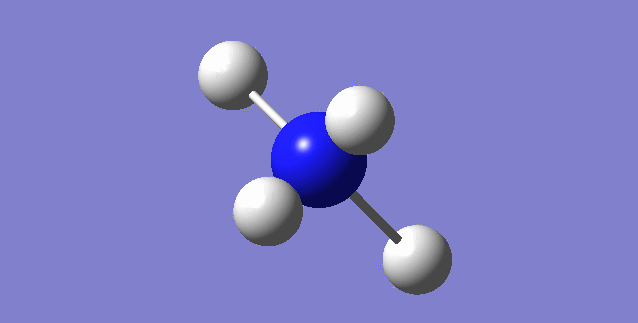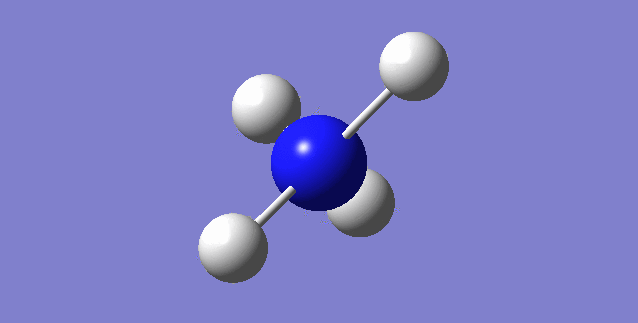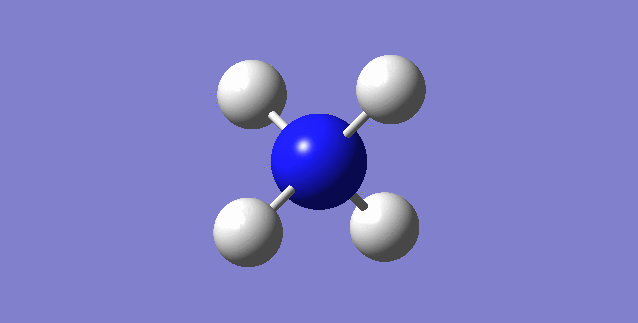
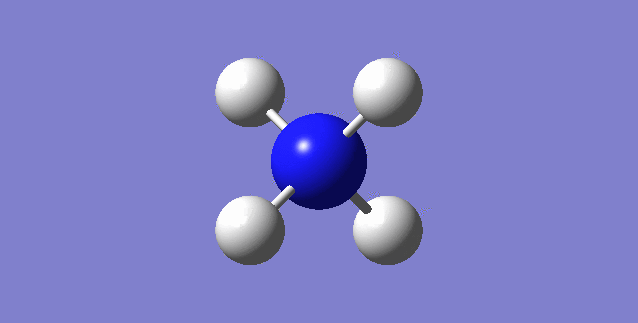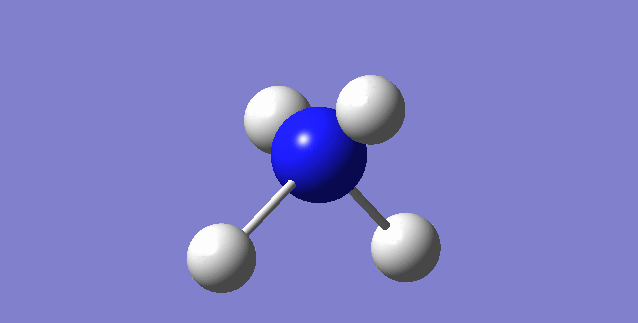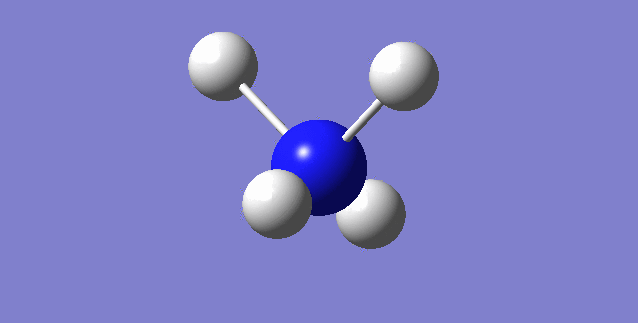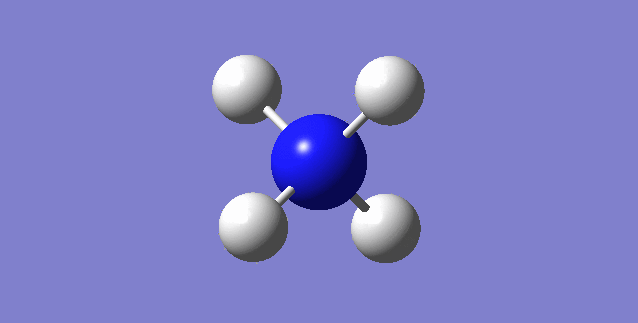
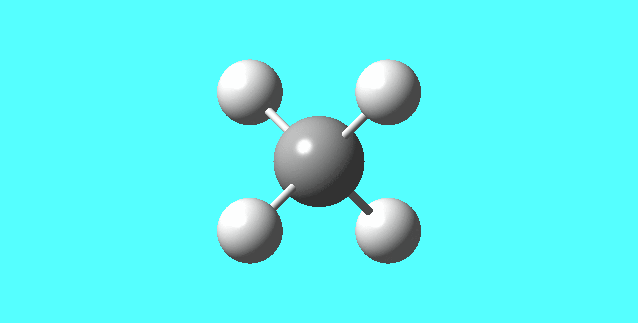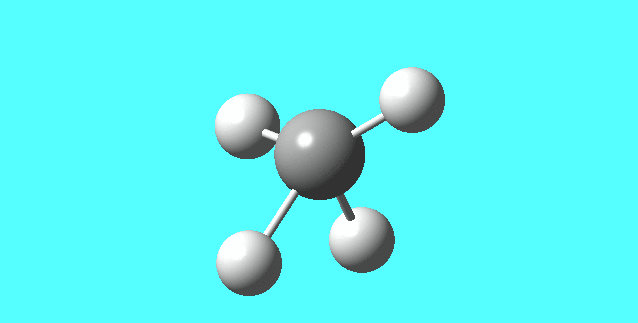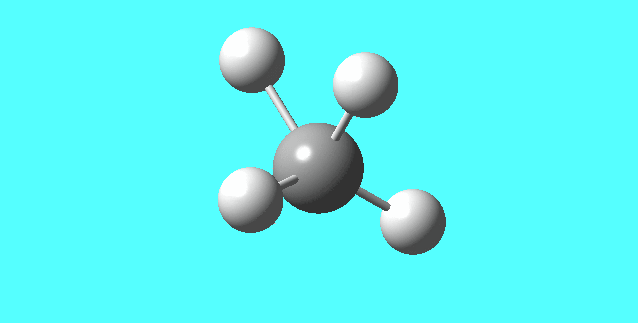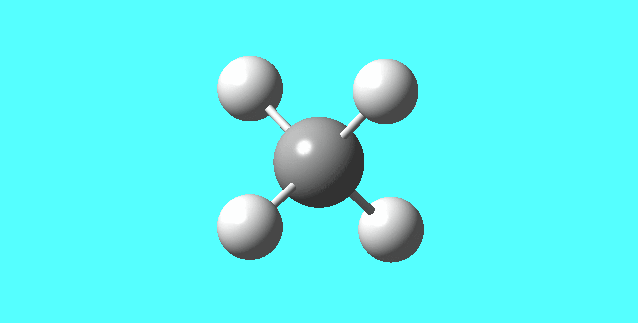
CH4
image of CH |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -40.52401404 a.u. |
| RMS Gradient Norm | 0.00003263a.u. |
| Point Group | TD |
| C-H bond length | 1.09197Å |
| H-C-H Angle | 109.47122° |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000063 | 0.000450 | YES |
| RMS | Force | 0.000034 | 0.000300 | YES |
| Maximum | Displacement | 0.000179 | 0.001800 | YES |
| RMS | Displacement | 0.000095 | 0.001200 | YES |
vibrations
| mode | Frequency | Infrared | IR active | type |
|---|---|---|---|---|
| 1 | 1356.20 | 14.1008 | yes | bending |
| 2 | 1356.20 | 14.1008 | yes | bending |
| 3 | 1356.20 | 14.1008 | yes | bending |
| 4 | 1578.58 | 0.0000 | no | bending |
| 5 | 1578.58 | 0.0000 | no | bending |
| 6 | 3046.46 | 0.0000 | no | stretching |
| 7 | 3162.33 | 25.3343 | yes | stretching |
| 8 | 3162.33 | 25.3343 | yes | stretching |
| 9 | 3162.33 | 25.3343 | yes | stretching |
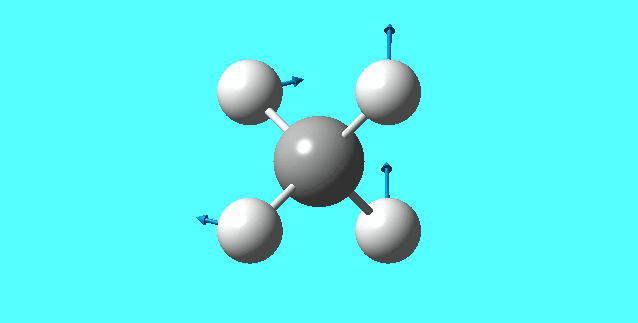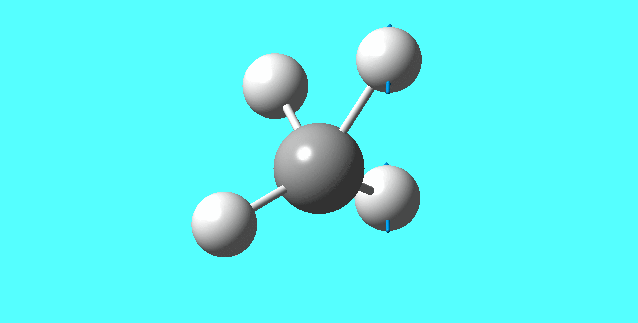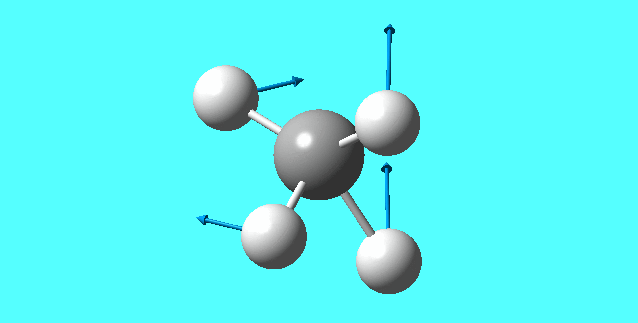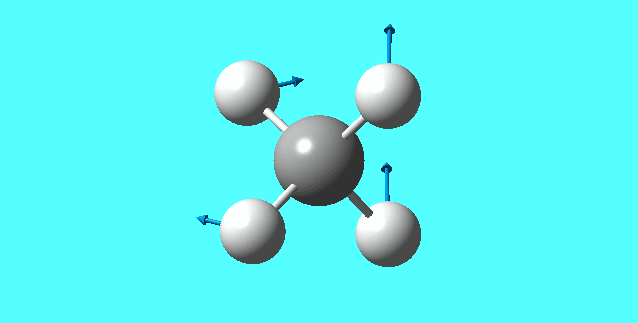
mode 1
mode 2
mode 3
mode 4
mode 5
mode 6
mode 7
mode 8
mode 9
Molecular orbitals
| MO | Occupancy | Energy (a.u.) |
|---|---|---|
| 15 | 0 | 0.8743736790 |
| 14 | 0 | 0.8743736790 |
| 13 | 0 | 0.8743736790 |
| 12 | 0 | 0.5291487570 |
| 11 | 0 | 0.5291487570 |
| 10 | 0 | 0.5291487570 |
| 9 | 0 | 0.1767699180 |
| 8 | 0 | 0.1767699180 |
| 7 | 0 | 0.1767699180 |
| 6 | 0 | 0.1182376210 |
| 5 | 2 | -0.3883090690 |
| 4 | 2 | -0.3883090690 |
| 3 | 2 | -0.3883090690 |
| 2 | 2 | -0.6904066760 |
| 1 | 2 | -10.1670726000 |
Molecular orbital diagrams
MO1

MO2

MO3-5

MO6

MO7-9

MO10 and above
these are imaginary orbitals and are a result of the linear fitting trying to produce results as can be seen for MO 15

data file
comparison
as the ammonium ion is isoelectronic to the methane it is similar to it in every way with it having the same number of peaks in the IR spectra however they would be at different locations to those for the methane. the MO also have different Energies with the charge going from negative to positive between the HOMO and LUMO for methane however it changes from MO9-MO10 which is above the LUMO due to if being electron deficient as it is a positively changed molecule.
- ↑ E. S. P. B. V, M.Sana,G.Leroy,D.Peeters,C.silante,D.C.Quantique and P.L.Pasteur,J. Mol. Struct. Theochem,1988,164,249-274