Es4215 march
NH3 molecule
Ammonia |
Optimisation data
Calculation type: RB3LYP
Basis set: 6-31G(d,p)
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000004 | 0.000450 | YES |
| RMS | Force | 0.000004 | 0.000300 | YES |
| Maximum | Displacement | 0.000072 | 0.001800 | YES |
| RMS | Displacement | 0.000035 | 0.001200 | YES |
Energy: -56.55776873 a.u.
RMS gradient: 0.00000485 a.u.
Point group: C3v
N-H bond length: 1.01798 Å
H-N-H bond angle: 105.741°
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
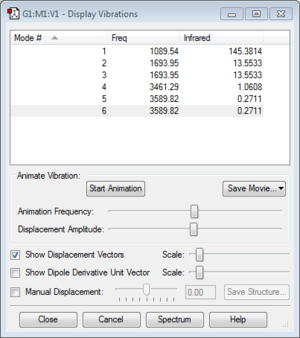
6 vibrational modes, as would be expected using the 3N-6 rule. Modes 2 and 3 are degenerate, as are modes 5 and 6, so 4 peaks would be expected in the IR spectrum for ammonia. Modes 1-3 are bending vibrations and 4-6 are bond stretch vibrations. Mode 4 is highly symmetric, but because of the trigonal pyramidal structure of ammonia, still has a shift in dipole moment and shows a peak on the experimental spectrum, though very weak[1]. Mode 1, a symmetrical bend of all three N-H bonds, is by far the most intense of the peaks because of a large shift in dipole moment and known as the "umbrella" mode.
The full optimisation file can be found here
Charge distribution
Charge on N atom: -1.125.
Charge on H atom: +0.375.
This is as expected, since the N atom is more electronegative than the H atom, so it would be expected to carry partial negative charge.
Thermodynamics of the Haber-Bosch process
The Haber-Bosch process is the process used to produce ammonia industrially, by reacting hydrogen and nitrogen directly together with an iron catalyst. It was first developed by Fritz Haber and Carl Bosch in the early 20th century. The stoichiometric reaction is:
N2 + 3H2 → 2NH3
The optimisation provided by Gaussview software can be used to analyse the thermodynamics of the reaction. In this case, the optimisation is being used to measure the internal energy of the reactants and products:
E(NH3)= -56.55776873 a.u.
2*E(NH3) = -113.11553746 a.u.
E(N2)= -109.52412828 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE= 2*E(NH3)-[E(N2)+3*E(H2)] = -0.0557911 a.u. = -146.47 kJ/mol
The forward reaction is exothermic. Ammonia is more stable and lower in energy than the elemental gases. This is in line with the conditions of the reaction, where decreasing the temperature favours the forward reaction according to Le Chatelier's principle. However, the experimentally determined value of the enthalpy is -92 kJ/mol[2], a difference of over 50 kJ/mol. Small discrepancies can be accounted for by taking into account the energy that is released as internal molecular motion, and energy that is absorbed by the walls of the container, but this discrepancy is large enough to suggest that there are processes happening in the reaction that are not accounted for by this method of calculation. Entropy changes could also have an effect: since the reaction converts 4 molecules to 2, there is a marked decrease in entropy.
N2
Optimisation data
Calculation method: RB3LYP
Basis set: 6-31(d,p)
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000001 | 0.000450 | YES |
| RMS | Force | 0.000001 | 0.000300 | YES |
| Maximum | Displacement | 0.000000 | 0.001800 | YES |
| RMS | Displacement | 0.000000 | 0.001200 | YES |
Energy: -109.52412828 a.u.
RMS gradient: 0.00000060 a.u.
Point group: D∞h
Bond length: 1.10550 Å
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
Frequency: 2457.33
Molecular Orbitals - occupied: 1σg, 1σ*u, 2sσg, 2sσ*u, 2pσg, 2pπu, 2pπu
- unoccupied: 2pπ*g, 2pπ*g, 2pσ*u and above
The full optimisation file can be found here
H2
Optimisation data
Calculation method: RB3LYP
Basis set: 6-31(d,p)
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000000 | 0.000450 | YES |
| RMS | Force | 0.000000 | 0.000300 | YES |
| Maximum | Displacement | 0.000000 | 0.001800 | YES |
| RMS | Displacement | 0.000001 | 0.001200 | YES |
Energy: -1.17853936 a.u.
RMS gradient: 0.00000017 a.u.
Point group: D∞h
Bond length: 0.74729 Å
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
Frequency: 4465.68
The full optimisation file can be found here
Project molecule ClF3
Optimisation data
Calculation method: RB3LYP
Basis set: 6-31(d,p)
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000050 | 0.000450 | YES |
| RMS | Force | 0.000028 | 0.000300 | YES |
| Maximum | Displacement | 0.000204 | 0.001800 | YES |
| RMS | Displacement | 0.000134 | 0.001200 | YES |
Energy: -759.46531688 a.u.
RMS gradient: 0.00002465 a.u.
Point group: C2v
The molecule is T-shaped, with a pseudostructure that is trigonal bipyramidal, as predicted by VSEPR (three bonding and two lone pairs). This results in the axial and equatorial F atoms being inequivalent.
Another possible structure of the molecule, a trigonal planar shape, was also analysed.
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000011 | 0.000450 | YES |
| RMS | Force | 0.000007 | 0.000300 | YES |
| Maximum | Displacement | 0.000049 | 0.001800 | YES |
| RMS | Displacement | 0.000032 | 0.001200 | YES |
The energy for this structure is -759.44149573 a.u., so this structure is higher in energy than the T-shape. Therefore, the T-shape is favoured.
Cl-F bond length (axial): 1.72863 Å
Cl-F bond length (equatorial): 1.65143 Å
F-Cl-F bond angle (axial-equatorial): 87.140°
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.7286 -DE/DX = 0.0 !
! R2 R(1,3) 1.6514 -DE/DX = 0.0 !
! R3 R(1,4) 1.7286 -DE/DX = 0.0 !
! A1 A(2,1,3) 87.1404 -DE/DX = 0.0 !
! A2 A(3,1,4) 87.1404 -DE/DX = 0.0 !
! A3 L(2,1,4,3,-1) 174.2807 -DE/DX = 0.0 !
! A4 L(2,1,4,3,-2) 180.0 -DE/DX = 0.0 !
! D1 D(2,1,3,4) 180.0 -DE/DX = 0.0 !
Frequency: 6 modes: 304.79, 309.07, 401.06, 540.78, 735.85, 752.26. Follows the 3N-6 rule.
The full optimisation file for the T-shaped structure can be found here and the file for the theoretical trigonal planar structure can be found here.
Charge distribution
Cl: +1.225
Axial F: -0.454
Equatorial F: -0.316
This is as expected as F is more electronegative than Cl, but it is interesting that the axial Cl-F bonds, which are longer, are more polar than the equatorial Cl-F bond.
Molecular orbitals
In ClF3, 22 molecular orbitals are occupied. The following are five of the various kinds of orbital interactions. It is interesting to note that more of the 22 MOs show bonding interactions between the Cl and axial Fs than between the Cl and the equatorial F, despite the fact that the axial Cl-F bonds are longer than the equatorial Cl-F bond. It is also interesting that none of the MOs obviously correspond to lone valence pairs on the Cl atom, as would be predicted by VSEPR This would suggest that whilst VSEPR is a useful tool for predicting molecule structures, it does not explain the actual electronic structure of the molecule.