Es4215 2nd yr Inorganic Comp
Part 1: BH3-NH3
BH3
BH3 |
First optimisation
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 3-21G
Charge = 0
Spin = Singlet
E(RB3LYP) = -26.46226371 a.u.
RMS Gradient Norm = 0.00008756 a.u.
Imaginary Freq =
Dipole Moment = 0.0003 Debye
Point Group = CS
Job cpu time: 0 days 0 hours 3 minutes 10.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000217 0.000450 YES
RMS Force 0.000105 0.000300 YES
Maximum Displacement 0.000692 0.001800 YES
RMS Displacement 0.000441 0.001200 YES
Predicted change in Energy=-1.635268D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1947 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1944 -DE/DX = -0.0001 !
! R3 R(1,4) 1.1948 -DE/DX = -0.0002 !
! A1 A(2,1,3) 119.9989 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0157 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9855 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Second optimisation
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -26.61532350 a.u.
RMS Gradient Norm = 0.00008174 a.u.
Imaginary Freq =
Dipole Moment = 0.0003 Debye
Point Group = CS
Job cpu time: 0 days 0 hours 3 minutes 19.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000203 0.000450 YES
RMS Force 0.000098 0.000300 YES
Maximum Displacement 0.000867 0.001800 YES
RMS Displacement 0.000415 0.001200 YES
Predicted change in Energy=-1.436134D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1926 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1924 -DE/DX = 0.0 !
! R3 R(1,4) 1.1928 -DE/DX = -0.0002 !
! A1 A(2,1,3) 119.999 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0146 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9864 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
After constraining the symmetry to D3H:
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -26.61532349 a.u.
RMS Gradient Norm = 0.00008043 a.u.
Imaginary Freq =
Dipole Moment = 0.0000 Debye
Point Group = D3H
Job cpu time: 0 days 0 hours 0 minutes 32.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000161 0.000450 YES
RMS Force 0.000105 0.000300 YES
Maximum Displacement 0.000638 0.001800 YES
RMS Displacement 0.000418 0.001200 YES
Predicted change in Energy=-1.539716D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1927 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1927 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1927 -DE/DX = -0.0002 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Log file of the final optimisation calculation here
Frequency calculation for the BH3 molecule
Calculation Type = FREQ
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -26.61532349 a.u.
RMS Gradient Norm = 0.00008047 a.u.
Imaginary Freq = 0
Dipole Moment = 0.0000 Debye
Point Group = D3H
Job cpu time: 0 days 0 hours 1 minutes 3.0 seconds.
Item Value Threshold Converged? Maximum Force 0.000161 0.000450 YES RMS Force 0.000080 0.000300 YES Maximum Displacement 0.000634 0.001800 YES RMS Displacement 0.000317 0.001200 YES Predicted change in Energy=-1.529731D-07 Optimization completed. -- Stationary point found.
Low frequencies --- -0.1187 -0.0051 -0.0013 42.2482 42.2484 43.3387 Low frequencies --- 1163.5889 1213.5519 1213.5521
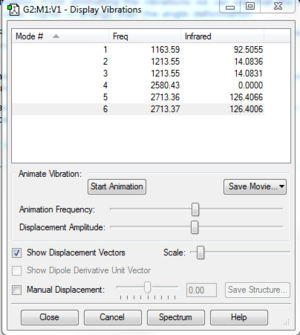
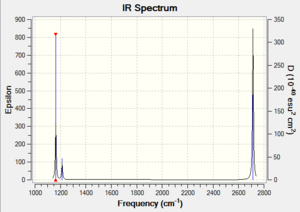
Ng611 (talk) 13:24, 6 June 2018 (BST) Good IR analysis. Regarding the MO diagram, how similar are the MOs predicted by qualitative MO to the calculated ones, and what does this tell you about the accuracy and usefullness of qualitative MO theory?
The three lower-frequency vibrations are bend vibrations whilst the higher-frequency vibrations are stretches. Vibration 4 is a symmetric stretch with no transition dipole moment and so it is not IR active. The absorption bands at 1213 and 2713 are each composed of two degenerate vibrational modes.
Log file of the frequency calculation here
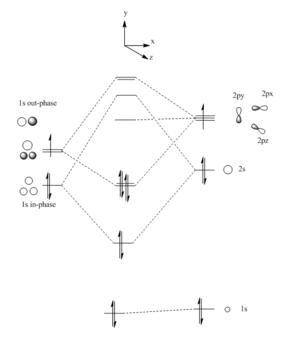
Lewis acid-base pair
Optimisation of NH3
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -56.55776870 a.u.
RMS Gradient Norm = 0.00011018 a.u.
Imaginary Freq =
Dipole Moment = 1.8460 Debye
Point Group = C1
Job cpu time: 0 days 0 hours 1 minutes 56.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000182 0.000450 YES
RMS Force 0.000093 0.000300 YES
Maximum Displacement 0.000235 0.001800 YES
RMS Displacement 0.000183 0.001200 YES
Predicted change in Energy=-7.297706D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0178 -DE/DX = 0.0002 !
! R2 R(1,3) 1.0182 -DE/DX = -0.0002 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7533 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7627 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7444 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.881 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Log file of the NH3 optimisation can be found here
Ng611 (talk) 13:28, 6 June 2018 (BST) You're missing a lot of data here. For every single calculation , we need: a screenshot of the summary table, the item table (which you've provided), the frequency log file (not the optimisation log file), the low-frequency modes, and a jmol of your final structure. You appear to have performed the calculations correctly, but without this data, it's impossible to confirm.
Optimisation of aminoborane
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -83.22468856 a.u.
RMS Gradient Norm = 0.00011867 a.u.
Imaginary Freq =
Dipole Moment = 5.5649 Debye
Point Group = C1
Job cpu time: 0 days 0 hours 3 minutes 23.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000279 0.000450 YES
RMS Force 0.000093 0.000300 YES
Maximum Displacement 0.000815 0.001800 YES
RMS Displacement 0.000435 0.001200 YES
Predicted change in Energy=-4.459583D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0188 -DE/DX = -0.0002 !
! R2 R(2,8) 1.0187 -DE/DX = -0.0002 !
! R3 R(3,8) 1.0185 -DE/DX = 0.0 !
! R4 R(4,7) 1.2097 -DE/DX = 0.0 !
! R5 R(5,7) 1.2104 -DE/DX = -0.0003 !
! R6 R(6,7) 1.2101 -DE/DX = -0.0002 !
! R7 R(7,8) 1.6684 -DE/DX = -0.0002 !
! A1 A(4,7,5) 113.8833 -DE/DX = 0.0 !
! A2 A(4,7,6) 113.893 -DE/DX = 0.0 !
! A3 A(4,7,8) 104.6021 -DE/DX = 0.0 !
! A4 A(5,7,6) 113.8649 -DE/DX = 0.0 !
! A5 A(5,7,8) 104.5766 -DE/DX = 0.0 !
! A6 A(6,7,8) 104.589 -DE/DX = 0.0 !
! A7 A(1,8,2) 107.8646 -DE/DX = 0.0 !
! A8 A(1,8,3) 107.8547 -DE/DX = 0.0 !
! A9 A(1,8,7) 111.0348 -DE/DX = 0.0 !
! A10 A(2,8,3) 107.8582 -DE/DX = 0.0 !
! A11 A(2,8,7) 111.0425 -DE/DX = 0.0 !
! A12 A(3,8,7) 111.0391 -DE/DX = 0.0 !
! D1 D(4,7,8,1) -180.0001 -DE/DX = 0.0 !
! D2 D(4,7,8,2) -59.9935 -DE/DX = 0.0 !
! D3 D(4,7,8,3) 60.008 -DE/DX = 0.0 !
! D4 D(5,7,8,1) -59.9967 -DE/DX = 0.0 !
! D5 D(5,7,8,2) 60.0099 -DE/DX = 0.0 !
! D6 D(5,7,8,3) -179.9887 -DE/DX = 0.0 !
! D7 D(6,7,8,1) 59.98 -DE/DX = 0.0 !
! D8 D(6,7,8,2) 179.9866 -DE/DX = 0.0 !
! D9 D(6,7,8,3) -60.0119 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Log file of the aminoborane optimisation can be found here
Final association calculation
Energy of BH3: -26.615323 a.u.
Energy of NH3: -56.557769 a.u.
Energy of aminoborane: -83.224689 a.u.
Energy change= -0.051678 a.u. = -135.68 kJ/mol
Ng611 (talk) 13:29, 6 June 2018 (BST) The calculation is performed correctly, but the final value should be reported to the nearest kj/mol.
BBr3
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = Gen
Charge = 0
Spin = Singlet
E(RB3LYP) = -64.43644947 a.u.
RMS Gradient Norm = 0.00000384 a.u.
Imaginary Freq =
Dipole Moment = 0.0000 Debye
Point Group = D3H
Job cpu time: 0 days 0 hours 0 minutes 31.6 seconds.
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.085961D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
A frequency analysis was then carried out.
Calculation Type = FREQ
Calculation Method = RB3LYP
Basis Set = Gen
Charge = 0
Spin = Singlet
E(RB3LYP) = -64.43644947 a.u.
RMS Gradient Norm = 0.00000392 a.u.
Imaginary Freq = 0
Dipole Moment = 0.0000 Debye
Point Group = D3H
Job cpu time: 0 days 0 hours 0 minutes 36.3 seconds.
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000037 0.001800 YES RMS Displacement 0.000018 0.001200 YES Predicted change in Energy=-4.301212D-10 Optimization completed. -- Stationary point found.
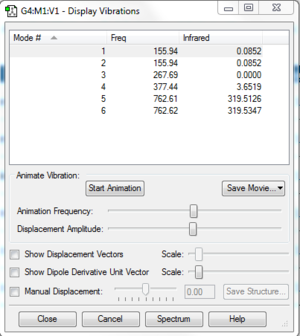
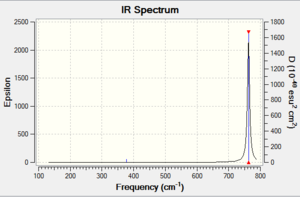
6 vibrational modes were found, including two sets of two degenerate modes each. One mode is IR inactive; one of the degenerate sets has intensity so small it does not appear on the spectrum. Another mode gives a small peak easy to overlook. Only one peak is easily noticed on the spectrum.
All the frequencies are lower than in the BH3 spectrum, possibly because Br is a heavier atom than B.
Also noticeable is the fact that the symmetric bend in BBr3 is higher in frequency relative to the other vibrational modes than in BH3.
Part 2: Project section-Lewis acids and bases
Al2Cl4Br2 exists as a Lewis acid-base pair with a number of different isomers:
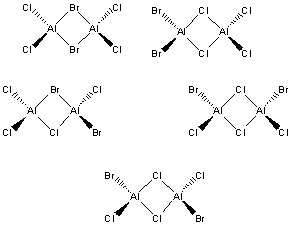
Isomer with both Br atoms at the same terminus. Projected point group C2v.
Isomer with both Br atoms bridging. Projected point group D2h.
Isomer with one Br atom bridging and the other terminal. Projected point group C1.
Isomer with both Br atoms at opposite termini, cis to one another. Projected point group C2v
Isomer with both Br atoms at opposite termini, trans to one another. Projected point group C2h
Ng611 (talk) 13:31, 6 June 2018 (BST) Good point group assignment!
Isomer with both Br atoms bridging
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Ng611 (talk) 13:38, 6 June 2018 (BST) Did you just use this basis set? For these calculations, you needed to use a mixed masis set (6-31G(d,p) for Al, Cl and LANL2DZ for Br).
Charge = 0
Spin = Singlet
E(RB3LYP) = -7469.54269098 a.u.
RMS Gradient Norm = 0.00004062 a.u.
Imaginary Freq =
Dipole Moment = 0.0007 Debye
Point Group = C1
Job cpu time: 0 days 0 hours 12 minutes 26.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000059 0.000450 YES
RMS Force 0.000027 0.000300 YES
Maximum Displacement 0.000842 0.001800 YES
RMS Displacement 0.000508 0.001200 YES
Predicted change in Energy=-1.390005D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,5) 2.0991 -DE/DX = 0.0 !
! R2 R(1,6) 2.4383 -DE/DX = 0.0001 !
! R3 R(1,7) 2.4383 -DE/DX = 0.0001 !
! R4 R(1,8) 2.0991 -DE/DX = 0.0 !
! R5 R(2,3) 2.0991 -DE/DX = 0.0 !
! R6 R(2,4) 2.0991 -DE/DX = 0.0 !
! R7 R(2,6) 2.4383 -DE/DX = 0.0001 !
! R8 R(2,7) 2.4383 -DE/DX = 0.0001 !
! A1 A(5,1,6) 110.2584 -DE/DX = 0.0 !
! A2 A(5,1,7) 110.2593 -DE/DX = 0.0 !
! A3 A(5,1,8) 120.2025 -DE/DX = 0.0 !
! A4 A(6,1,7) 92.0028 -DE/DX = 0.0 !
! A5 A(6,1,8) 110.2587 -DE/DX = 0.0 !
! A6 A(7,1,8) 110.2577 -DE/DX = 0.0 !
! A7 A(3,2,4) 120.2025 -DE/DX = 0.0 !
! A8 A(3,2,6) 110.259 -DE/DX = 0.0 !
! A9 A(3,2,7) 110.2586 -DE/DX = 0.0 !
! A10 A(4,2,6) 110.2581 -DE/DX = 0.0 !
! A11 A(4,2,7) 110.2585 -DE/DX = 0.0 !
! A12 A(6,2,7) 92.0028 -DE/DX = 0.0 !
! A13 A(1,6,2) 87.9972 -DE/DX = 0.0 !
! A14 A(1,7,2) 87.9971 -DE/DX = 0.0 !
! D1 D(5,1,6,2) 112.4854 -DE/DX = 0.0 !
! D2 D(7,1,6,2) 0.0133 -DE/DX = 0.0 !
! D3 D(8,1,6,2) -112.4572 -DE/DX = 0.0 !
! D4 D(5,1,7,2) -112.4846 -DE/DX = 0.0 !
! D5 D(6,1,7,2) -0.0133 -DE/DX = 0.0 !
! D6 D(8,1,7,2) 112.4581 -DE/DX = 0.0 !
! D7 D(3,2,6,1) 112.4582 -DE/DX = 0.0 !
! D8 D(4,2,6,1) -112.4845 -DE/DX = 0.0 !
! D9 D(7,2,6,1) -0.0133 -DE/DX = 0.0 !
! D10 D(3,2,7,1) -112.4585 -DE/DX = 0.0 !
! D11 D(4,2,7,1) 112.4841 -DE/DX = 0.0 !
! D12 D(6,2,7,1) 0.0133 -DE/DX = 0.0 !
-------------------------------------------------------------
Al-Br bond length: 2.438 Å. All bonds in the Al-Br-Al-Br bridging system were of equal length.
Al-Br-Al bridging bond angle: 87.997o
Al-Cl bond length: 2.099 Å
Full log file can be found here
Isomer with both Br atoms terminal and trans
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -7469.53761798 a.u.
RMS Gradient Norm = 0.00000456 a.u.
Imaginary Freq =
Dipole Moment = 0.0003 Debye
Point Group = C1
Job cpu time: 0 days 0 hours 13 minutes 44.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000007 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000108 0.001800 YES
RMS Displacement 0.000050 0.001200 YES
Predicted change in Energy=-1.700479D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,4) 2.0968 -DE/DX = 0.0 !
! R2 R(1,5) 2.2975 -DE/DX = 0.0 !
! R3 R(1,6) 2.2976 -DE/DX = 0.0 !
! R4 R(1,8) 2.2285 -DE/DX = 0.0 !
! R5 R(2,3) 2.0968 -DE/DX = 0.0 !
! R6 R(2,5) 2.2976 -DE/DX = 0.0 !
! R7 R(2,6) 2.2975 -DE/DX = 0.0 !
! R8 R(2,7) 2.2285 -DE/DX = 0.0 !
! A1 A(4,1,5) 108.9637 -DE/DX = 0.0 !
! A2 A(4,1,6) 108.9617 -DE/DX = 0.0 !
! A3 A(4,1,8) 122.7391 -DE/DX = 0.0 !
! A4 A(5,1,6) 90.7293 -DE/DX = 0.0 !
! A5 A(5,1,8) 110.3804 -DE/DX = 0.0 !
! A6 A(6,1,8) 110.3783 -DE/DX = 0.0 !
! A7 A(3,2,5) 108.9619 -DE/DX = 0.0 !
! A8 A(3,2,6) 108.9642 -DE/DX = 0.0 !
! A9 A(3,2,7) 122.7396 -DE/DX = 0.0 !
! A10 A(5,2,6) 90.7286 -DE/DX = 0.0 !
! A11 A(5,2,7) 110.3778 -DE/DX = 0.0 !
! A12 A(6,2,7) 110.3799 -DE/DX = 0.0 !
! A13 A(1,5,2) 89.2707 -DE/DX = 0.0 !
! A14 A(1,6,2) 89.2714 -DE/DX = 0.0 !
! D1 D(4,1,5,2) 110.3536 -DE/DX = 0.0 !
! D2 D(6,1,5,2) -0.0106 -DE/DX = 0.0 !
! D3 D(8,1,5,2) -112.1109 -DE/DX = 0.0 !
! D4 D(4,1,6,2) -110.3555 -DE/DX = 0.0 !
! D5 D(5,1,6,2) 0.0106 -DE/DX = 0.0 !
! D6 D(8,1,6,2) 112.1128 -DE/DX = 0.0 !
! D7 D(3,2,5,1) 110.3769 -DE/DX = 0.0 !
! D8 D(6,2,5,1) 0.0106 -DE/DX = 0.0 !
! D9 D(7,2,5,1) -112.0908 -DE/DX = 0.0 !
! D10 D(3,2,6,1) -110.3748 -DE/DX = 0.0 !
! D11 D(5,2,6,1) -0.0106 -DE/DX = 0.0 !
! D12 D(7,2,6,1) 112.0888 -DE/DX = 0.0 !
-------------------------------------------------------------
Al-Br bond length: 2.228 Å
Al-Cl bond length (bridging): 2.2975 Å and 2.2976 Å. Opposing sides of the Cl-Al-Cl-Al bridging system were equal in length. The two different Al-Cl bond lengths reduced the symmetry to C1 instead of the expected C2h.
Al-Cl bond length (terminal): 2.097 Å
Al-Cl-Al bridging bond angle: 89.271o. A larger angle than the Al-Br-Al bridging angle.
Full log file for this computation can be found here
The energy calculated for this isomer is -7469.537618 a.u., in comparison to the bridging isomer, whose energy is -7469.542691 a.u.. Therefore the bridging isomer is lower in energy by 0.005073 a.u., which corresponds to an interconversion energy of ±13.3 kJ/mol.
The molecule is more stable with Br as the bridging atoms. A plausible explanation is that Br is a larger atom than Cl, and therefore the four-atom bridging system is less strained. This is supported by several pieces of evidence from the Gaussian calculations. Firstly, there are two different Al-Cl bridging bond lengths in the trans isomer, suggesting a more strained system. In comparison, the Al-Br bonds in the bridging isomer are all equal. Al-Cl bridging bonds are also shorter than Al-Br bridging bonds, and the Al-Cl-Al bridging angle is larger than the Al-Br-Al angle. All of these indicate that Al-Cl-Al-Cl bridging system is more strained than the Al-Br-Al-Br bridging system.
Association energy of the bridging isomer
The bridging isomer is lower in energy and therefore the more stable of the two isomers. Therefore, since the isomers are able to interconvert around the Al atoms, the bridging isomer will be preferred. It can be formed by dimerisation of AlCl2Br, which is both Lewis acidic (as it is electron deficient around the Al atom) and has Lewis base character (due to lone pairs of electrons on the halogens). In order to calculate the association energy of this process, optimisation of the monomer AlCl2Br can be carried out.
Calculation Type = FOPT
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -3734.74851982 a.u.
RMS Gradient Norm = 0.00001669 a.u.
Imaginary Freq =
Dipole Moment = 0.6912 Debye
Point Group = C2V
Job cpu time: 0 days 0 hours 8 minutes 44.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000014 0.000300 YES
Maximum Displacement 0.000164 0.001800 YES
RMS Displacement 0.000105 0.001200 YES
Predicted change in Energy=-5.066756D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.2235 -DE/DX = 0.0 !
! R2 R(1,3) 2.0912 -DE/DX = 0.0 !
! R3 R(1,4) 2.0912 -DE/DX = 0.0 !
! A1 A(2,1,3) 121.075 -DE/DX = 0.0 !
! A2 A(2,1,4) 121.075 -DE/DX = 0.0 !
! A3 A(3,1,4) 117.8499 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
-------------------------------------------------------------
Full log file for this optimisation can be found here
Energy of the Br-bridging dimer: -7469.542618 a.u.
Energy of AlCl2Br: -3734.748520 a.u.
Energy change at association: -0.045778 a.u.=-119.7 kJ/mol
Molecular orbitals of the bridging isomer
bridging Al2Cl4Br2 |
To calculate the molecular orbitals of the bridging isomer, a frequency analysis of the bridging isomer was carried out.
Calculation Type = FREQ
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
Charge = 0
Spin = Singlet
E(RB3LYP) = -7469.54269097 a.u.
RMS Gradient Norm = 0.00004063 a.u.
Imaginary Freq = 0
Dipole Moment = 0.0007 Debye
Point Group = C1
Job cpu time: 0 days 0 hours 2 minutes 31.0 seconds.
Item Value Threshold Converged? Maximum Force 0.000097 0.000450 YES RMS Force 0.000041 0.000300 YES Maximum Displacement 0.001672 0.001800 YES RMS Displacement 0.000687 0.001200 YES Predicted change in Energy=-1.760083D-07 Optimization completed. -- Stationary point found.
Full log file of the frequency computation can be found here
| MO number | Visualised MO | Diagram of constituent atomic orbitals | Description |
|---|---|---|---|
| 72 | 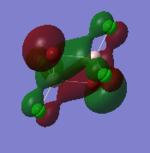 |
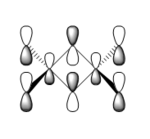 |
This orbital is composed of pz orbitals, all locally in phase and thus able to interact in a bonding fashion. There is a large area of electron density between the Al atoms and the bridging Br atoms, indicating a σ-style overlap and thus a strong bonding interaction. There are bonding interactions through-bond between the Al atoms and the terminal Cl atoms, but these are π-style and thus weaker. Through-space bonding interactions between the terminal Cl atoms are possible, but very weak because of the distance being larger and the interactions being π-type. Since Al is less electronegative than either Cl and Br, the Al contribution to this bonding orbital is smaller. Overall: a highly bonding orbital. |
| 79 | 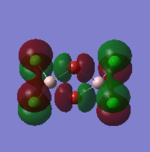 |
 |
This orbital is also composed of p-orbitals. The p-orbitals on the Cl atoms at the same terminus are in phase, with a very weak π-type through-space bonding interaction. Between Cl atoms on opposite termini, there are extremely weak π-type through-space antibonding interactions. The p-orbitals on the bridging Br atoms are oriented orthogonally to those on the Cl atoms. Since they are oriented closer to the opposite-phase lobe of the p-orbital on the Cl atoms, the antibonding interactions will be slightly stronger. There are also weak π-type antibonding interactions between the p-orbitals on the Br atoms. The Al atoms do not contribute to this MO. Overall: non-bonding, maybe slightly antibonding. Ng611 (talk) 13:41, 6 June 2018 (BST) You have to choose one or the other (It is a tough one to call to be fair). |
| 81 | 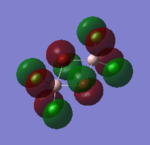 |
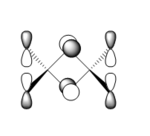 |
This orbital is also composed of p-orbitals. There are weak π-type through-space antibonding interactions between orbitals on Cl atoms at the same terminus, and between the orbitals on the bridging Br atoms. Stronger σ-type through-space antibonding interactions occur between orbitals on the bridging Br atoms and terminal Cl atoms. There are very weak π-type through-space interactions between orbitals on Cl atoms at opposite termini. As with Molecular Orbital 79, the Al atoms do not contribute to this MO. Overall: highly antibonding. |
Ng611 (talk) 13:38, 6 June 2018 (BST) Good LCAO decompositions, and a good description of the main interactions (although annotating your LCAO diagrams as Tricia has done in her lectures would be better).
Ng611 (talk) 13:46, 6 June 2018 (BST) Overall, some really good parts to this report. You LCAO analysis, MO diagram, and point group analysis were performed excellently. You let yourself down by not including the relevant information for your calculations, meaning that there was no way for us to assess how well you performed them -- with this information, you would have had a pretty good report.
