EX3 section BC2116
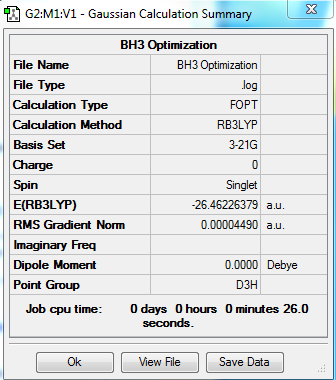
Boron Hydride (BH3)
B3LYP/3-21G level
Summary table:
Item table:
Item Value Threshold Converged?
Maximum Force 0.000203 0.000450 YES
RMS Force 0.000098 0.000300 YES
Maximum Displacement 0.000867 0.001800 YES
RMS Displacement 0.000415 0.001200 YES
Predicted change in Energy=-1.436186D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1928 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1926 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1924 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0146 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9864 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.999 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
log file:
3D model:
test molecule |
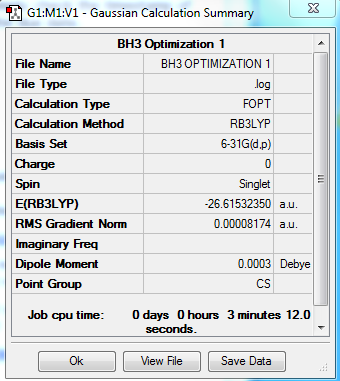
B3LYP/6-31G level
Summary table:
Item table:
Item Value Threshold Converged?
Maximum Force 0.000203 0.000450 YES
RMS Force 0.000098 0.000300 YES
Maximum Displacement 0.000867 0.001800 YES
RMS Displacement 0.000415 0.001200 YES
Predicted change in Energy=-1.436134D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1928 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1926 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1924 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0146 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9864 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.999 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
log file:
3D model:
test molecule |
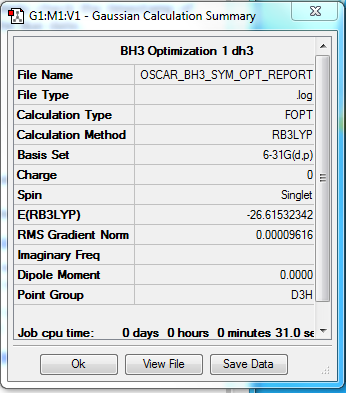
B3LYP/6-31G level D3H Optimization
Summary table:
Item table:
Item Value Threshold Converged?
Maximum Force 0.000192 0.000450 YES
RMS Force 0.000126 0.000300 YES
Maximum Displacement 0.000763 0.001800 YES
RMS Displacement 0.000500 0.001200 YES
Predicted change in Energy=-2.201121D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1927 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1927 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1927 -DE/DX = -0.0002 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
log file:
File:OSCAR BH3 SYM OPT REPORT.LOG
3D model:
test molecule |
Ng611 (talk) 23:17, 15 May 2018 (BST) It's good that you're trying to be thorough by including the results of your pre optimisation calculation, but we don't need to see this, just the results of your final calculation.
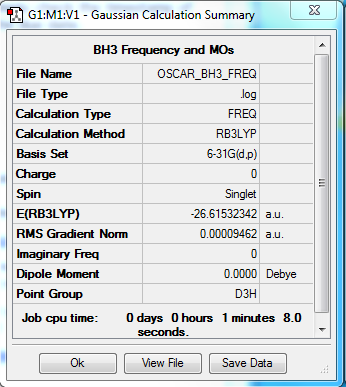
B3LYP/6-31G level frequency calculation
Summary table:
Item table:
Item Value Threshold Converged?
Maximum Force 0.000189 0.000450 YES
RMS Force 0.000095 0.000300 YES
Maximum Displacement 0.000746 0.001800 YES
RMS Displacement 0.000373 0.001200 YES
Predicted change in Energy=-2.116143D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
low frequencies
Low frequencies --- 1163.7209 1213.6704 1213.6731
Diagonal vibrational polarizability:
0.7197914 0.7196912 1.8376208
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2" E' E'
Frequencies -- 1163.7209 1213.6704 1213.6731
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9998 0.9609 0.9609
IR Inten -- 92.4742 14.0889 14.0925
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
4 5 6
Ng611 (talk) 21:04, 17 May 2018 (BST) There should be two lines of frequency data reported -- one line for low frequencies close to and potentially less than 0 and another for 'higher' low frequencies. You've included the latter of the two above, but not the former.
log file:
3D model:
test molecule |
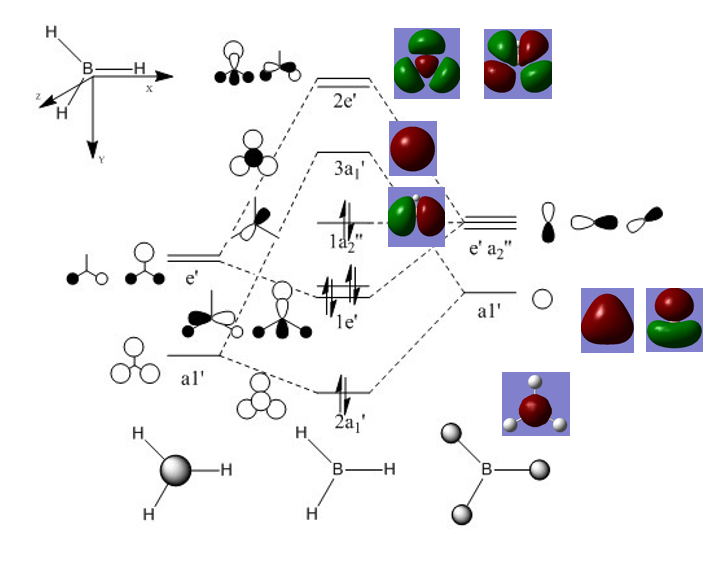
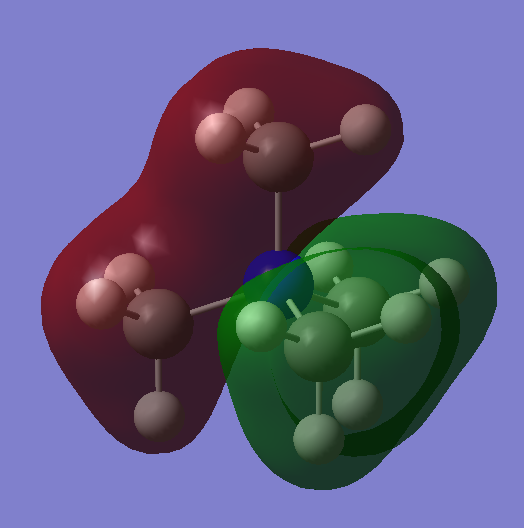
BH3 Molecular Orbitals (LCAO and Gaussian)
Significant difference between Gaussview and LCAO molecular orbitals, Gaussian orbitals seemed to be in the wrong order. This shows that although Gaussian program could produce good prediction of MO shapes, however sometimes it would fail to order them correctly. This was not an isolated incidence, other people had also encountered the same issue with their programs, and each error produced different orders.
BH3NH3 reaction enthalpy calculation
BH3NH3 summary table
Item table
Item Value Threshold Converged?
Maximum Force 0.000233 0.000450 YES
RMS Force 0.000083 0.000300 YES
Maximum Displacement 0.001351 0.001800 YES
RMS Displacement 0.000417 0.001200 YES
Predicted change in Energy=-5.216895D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0184 -DE/DX = 0.0 !
! R2 R(2,7) 1.0184 -DE/DX = 0.0 !
! R3 R(3,7) 1.0184 -DE/DX = 0.0 !
! R4 R(4,8) 1.2103 -DE/DX = -0.0002 !
! R5 R(5,8) 1.2103 -DE/DX = -0.0002 !
! R6 R(6,8) 1.2103 -DE/DX = -0.0002 !
! R7 R(7,8) 1.6671 -DE/DX = 0.0001 !
! A1 A(1,7,2) 107.9016 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.9016 -DE/DX = 0.0 !
! A3 A(1,7,8) 110.9986 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.9016 -DE/DX = 0.0 !
! A5 A(2,7,8) 110.9986 -DE/DX = 0.0 !
! A6 A(3,7,8) 110.9986 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.857 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.857 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.6185 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.857 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.6185 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.6185 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.9824 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -60.0176 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 59.9824 -DE/DX = 0.0 !
! D4 D(2,7,8,4) 59.9824 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 179.9824 -DE/DX = 0.0 !
! D6 D(2,7,8,6) -60.0176 -DE/DX = 0.0 !
! D7 D(3,7,8,4) -60.0176 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 59.9824 -DE/DX = 0.0 !
! D9 D(3,7,8,6) 179.9824 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low Frequencies
Low frequencies --- -0.0641 -0.0630 -0.0073 16.6819 16.6931 41.4307
Low frequencies --- 265.4200 634.4478 639.9302
Diagonal vibrational polarizability:
2.5473263 2.5473375 5.0085052
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A E
Frequencies -- 265.4074 634.4478 639.9299
Red. masses -- 1.0078 4.9892 1.0452
Frc consts -- 0.0418 1.1832 0.2522
IR Inten -- 0.0000 13.8547 3.5302
Atom AN X Y Z X Y Z X Y Z
1 1 0.00 -0.45 0.00 0.00 0.00 0.36 0.00 0.21 0.00
2 1 -0.39 0.22 0.00 0.00 0.00 0.36 -0.02 0.18 -0.51
3 1 0.39 0.22 0.00 0.00 0.00 0.36 0.02 0.18 0.51
4 1 0.00 -0.36 0.00 0.03 0.00 -0.29 0.00 0.15 0.00
5 1 -0.32 0.18 0.00 -0.02 -0.03 -0.29 -0.02 0.12 -0.40
6 1 0.32 0.18 0.00 -0.02 0.03 -0.29 0.02 0.12 0.40
7 7 0.00 0.00 0.00 0.00 0.00 0.36 0.00 -0.05 0.00
8 5 0.00 0.00 0.00 0.00 0.00 -0.48 0.00 -0.03 0.00
4 5 6
log file
3D Model
test molecule |
Energy Calculation
E(NH3)= -56.55776863 E(BH3)= -26.61532342 E(NH3BH3)= -83.22468997
ΔE = E(NH3BH3)-[E(NH3)+E(BH3)]
= -0.05159792 au
= -135.470339 kJ/mol
Ng611 (talk) 23:19, 15 May 2018 (BST) The final value should be reported to the nearest kj/mol
This is a relatively weak bond in comparison to C-C dative bond (350 kJ/mol).
Ng611 (talk) 23:19, 15 May 2018 (BST) Remember to cite your bond values (ideally from a textbook, databook, or paper).
BBr3
Summary
Item Table
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027258D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies
Low frequencies --- -0.0136 -0.0064 -0.0046 2.4322 2.4323 4.8425
Low frequencies --- 155.9631 155.9651 267.7051
Diagonal vibrational polarizability:
14.8692985 14.8689584 0.6892246
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E' E' A1'
Frequencies -- 155.9631 155.9651 267.7051
Red. masses -- 68.4421 68.4435 78.9183
Frc consts -- 0.9809 0.9809 3.3323
IR Inten -- 0.0843 0.0843 0.0000
Atom AN X Y Z X Y Z X Y Z
1 5 -0.39 0.00 0.00 0.00 0.39 0.00 0.00 0.00 0.00
2 35 0.55 0.00 0.00 0.00 0.51 0.00 0.00 0.58 0.00
3 35 -0.25 -0.46 0.00 0.46 -0.28 0.00 -0.50 -0.29 0.00
4 35 -0.25 0.46 0.00 -0.46 -0.28 0.00 0.50 -0.29 0.00
4 5 6
DOI
My DOI is: DOI:10042/202350
Benzene and Borazine
Comparison in calculated data
Summary
Item table of benzene and borazine
Benzene
Item Value Threshold Converged?
Maximum Force 0.000193 0.000450 YES
RMS Force 0.000094 0.000300 YES
Maximum Displacement 0.000787 0.001800 YES
RMS Displacement 0.000368 0.001200 YES
Predicted change in Energy=-4.828252D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Borazine
Item Value Threshold Converged?
Maximum Force 0.000199 0.000450 YES
RMS Force 0.000063 0.000300 YES
Maximum Displacement 0.000302 0.001800 YES
RMS Displacement 0.000100 0.001200 YES
Predicted change in Energy=-1.063835D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies of benzene and borazine
Benzene:
Low frequencies --- 414.0392 414.6031 621.0860
Diagonal vibrational polarizability:
0.2796010 0.2794350 4.1548187
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 414.0392 414.6031 621.0860
Red. masses -- 2.9400 2.9439 6.0699
Frc consts -- 0.2970 0.2982 1.3795
IR Inten -- 0.0001 0.0000 0.0000
Atom AN X Y Z X Y Z X Y Z
1 6 0.00 0.00 -0.12 0.00 0.00 0.21 0.34 -0.06 0.00
2 6 0.00 0.00 0.24 0.00 0.00 0.00 0.04 0.28 0.00
3 6 0.00 0.00 -0.12 0.00 0.00 -0.21 -0.04 0.16 0.00
4 6 0.00 0.00 -0.12 0.00 0.00 0.21 -0.34 0.06 0.00
5 6 0.00 0.00 0.24 0.00 0.00 0.00 -0.04 -0.28 0.00
6 6 0.00 0.00 -0.12 0.00 0.00 -0.21 0.04 -0.16 0.00
7 1 0.00 0.00 -0.26 0.00 0.00 0.45 0.31 -0.16 0.00
8 1 0.00 0.00 0.52 0.00 0.00 0.00 -0.21 0.22 0.00
9 1 0.00 0.00 -0.26 0.00 0.00 -0.45 0.21 -0.11 0.00
10 1 0.00 0.00 -0.26 0.00 0.00 0.45 -0.31 0.16 0.00
11 1 0.00 0.00 0.52 0.00 0.00 0.00 0.21 -0.22 0.00
12 1 0.00 0.00 -0.26 0.00 0.00 -0.45 -0.21 0.11 0.00
4 5 6
Borazine:
Low frequencies --- -6.6257 -6.4464 -5.9950 -0.0097 0.0573 0.1401
Low frequencies --- 289.2556 289.2660 403.8558
Diagonal vibrational polarizability:
7.3617472 7.3609937 14.1821308
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E" E" A2"
Frequencies -- 289.2556 289.2660 403.8558
Red. masses -- 2.9261 2.9262 1.9254
Frc consts -- 0.1442 0.1443 0.1850
IR Inten -- 0.0000 0.0000 23.7922
Atom AN X Y Z X Y Z X Y Z
1 1 0.00 0.00 0.26 0.00 0.00 -0.07 0.00 0.00 0.16
2 1 0.00 0.00 -0.18 0.00 0.00 0.67 0.00 0.00 0.53
3 1 0.00 0.00 -0.19 0.00 0.00 -0.19 0.00 0.00 0.16
4 1 0.00 0.00 0.67 0.00 0.00 -0.18 0.00 0.00 0.53
5 1 0.00 0.00 -0.07 0.00 0.00 0.26 0.00 0.00 0.16
6 1 0.00 0.00 -0.49 0.00 0.00 -0.49 0.00 0.00 0.53
7 5 0.00 0.00 0.22 0.00 0.00 -0.06 0.00 0.00 0.10
8 5 0.00 0.00 -0.06 0.00 0.00 0.22 0.00 0.00 0.10
9 5 0.00 0.00 -0.16 0.00 0.00 -0.16 0.00 0.00 0.10
10 7 0.00 0.00 -0.06 0.00 0.00 0.24 0.00 0.00 -0.13
11 7 0.00 0.00 -0.17 0.00 0.00 -0.17 0.00 0.00 -0.13
12 7 0.00 0.00 0.24 0.00 0.00 -0.06 0.00 0.00 -0.13
4 5 6
log files
File:OSCAR BENZENE OPT.LOG File:OSCAR BORAZINE OPT.LOG
3D Model
test molecule |
test molecule |
Analysis
Charge Distribution
Benzene and borazine have different charge distribution despite similarity in structure. The differences are tabulated below:
| Benzene | Borazine | |
|---|---|---|
| Boron | n/a | 0.074 |
| Carbon | -2.390 | n/a |
| Nitrogen | n/a | -1.102 |
| Hydrogen | 0.235 | 0.432 |
| Hydrogen | n/a | 0.077 |
Borazine atoms have four different electron densities, whereas benzene atoms only have two. This is because of the difference in electronegativity between nitrogen and boron. As nitrogen is more electronegative, the hydrogen bonded to them will have lower electron density. This electronegativity causes polarization in electron density, and decrease the delocalization of p orbital electrons. The decrease in delocalization causes resonance effect in borazine to be weaker than that in benzene. This is can be reflected in their chemical data, where benzene is lower in energy comparing to borazine