ERHW DFT calc
Appearance
Level 1 Lab classes: DFT calculations
Practice simulations and optimisations
NH3 optimisation data
Method: B3LYP
Basis set: 6-31G(d,p)
Calculations: OPTF
Point group: C3v
Optimised NH3 molecule
test molecule |
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986279D-10
Bond distances and bond angles ! R1 R(1,2) 1.018 -DE/DX = 0.0 ! ! R2 R(1,3) 1.018 -DE/DX = 0.0 ! ! R3 R(1,4) 1.018 -DE/DX = 0.0 ! ! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 ! ! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 ! ! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 ! ! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
XYZ file
4 NH<sub>3</sub>molecule N 0.0000 0.0000 0.1192 N H 0.0000 0.9372 -0.2782 H H -0.8116 -0.4686 -0.2782 H H 0.8116 -0.4686 -0.2782 H
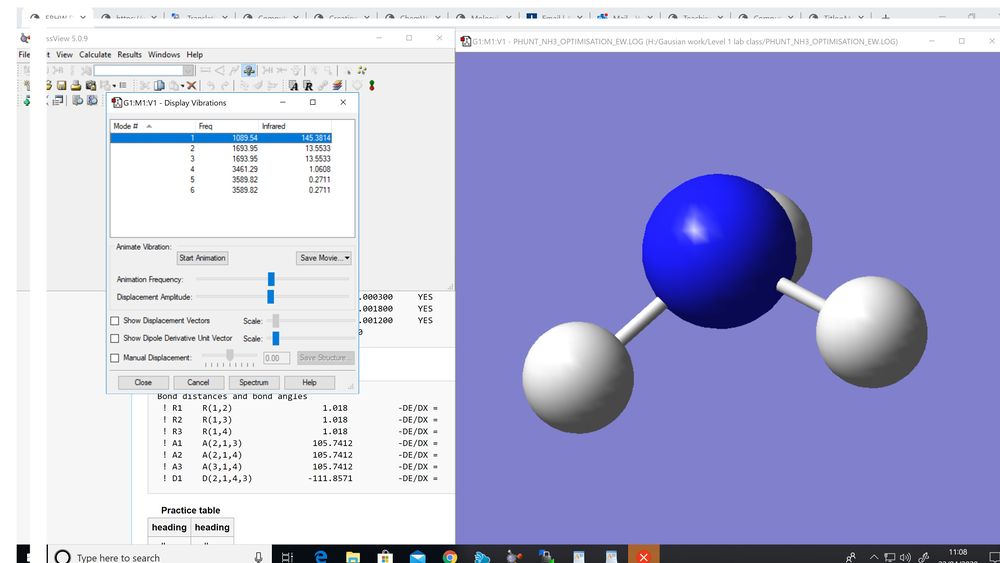
Vibrational modes
| 1 | 2 | 3 | 4 | 5 | 6 | ||
|---|---|---|---|---|---|---|---|
| wavenumber (cm-1) | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 | |
| symmetry | A1 | E | E | A1 | E | E | |
| Intensity (arbitrary units) | 145 | 14 | 14 | 1 | 0.3 | 0.3 | |
| Diagram | 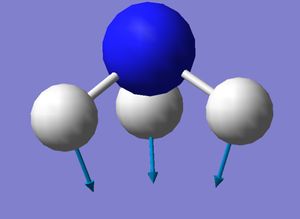 |
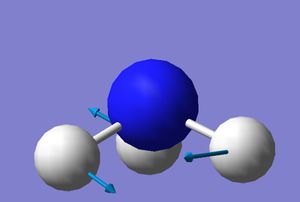 |
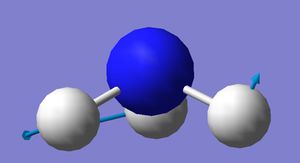
|
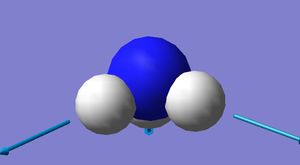 |
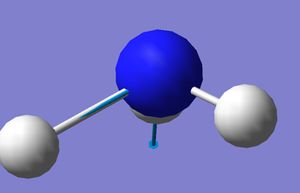 |
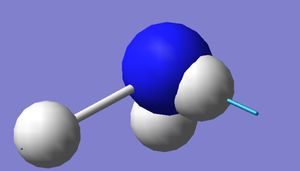 |
Charge distribution
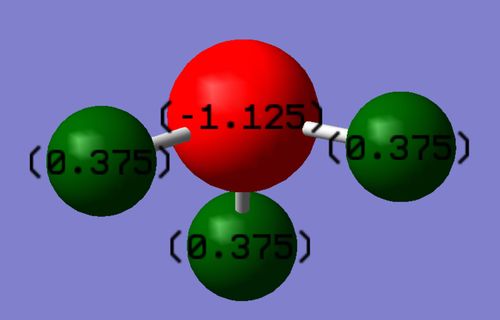
PF5optimisation data
Method: B3LYP
Basis set: 6-31G(d,p)
Calculations: OPTF
Point group: D3h
test molecule |
Item Value Threshold Converged? Maximum Force 0.000027 0.000450 YES RMS Force 0.000011 0.000300 YES Maximum Displacement 0.000061 0.001800 YES RMS Displacement 0.000026 0.001200 YES Predicted change in Energy=-3.393566D-09
Bond distances and bond angles ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 1.5693 -DE/DX = 0.0 ! ! R2 R(1,3) 1.597 -DE/DX = 0.0 ! ! R3 R(1,4) 1.5693 -DE/DX = 0.0 ! ! R4 R(1,5) 1.5693 -DE/DX = 0.0 ! ! R5 R(1,6) 1.597 -DE/DX = 0.0 ! ! A1 A(2,1,3) 90.0 -DE/DX = 0.0 ! ! A2 A(2,1,4) 120.0 -DE/DX = 0.0 ! ! A3 A(2,1,5) 120.0 -DE/DX = 0.0 ! ! A4 A(2,1,6) 90.0 -DE/DX = 0.0 ! ! A5 A(3,1,4) 90.0 -DE/DX = 0.0 ! ! A6 A(3,1,5) 90.0 -DE/DX = 0.0 ! ! A7 A(4,1,5) 120.0 -DE/DX = 0.0 ! ! A8 A(4,1,6) 90.0 -DE/DX = 0.0 ! ! A9 A(5,1,6) 90.0 -DE/DX = 0.0 ! ! A10 L(3,1,6,2,-1) 180.0 -DE/DX = 0.0 ! ! A11 L(3,1,6,2,-2) 180.0 -DE/DX = 0.0 ! ! D1 D(2,1,4,3) -90.0 -DE/DX = 0.0 ! ! D2 D(2,1,5,3) 90.0 -DE/DX = 0.0 ! ! D3 D(2,1,5,4) 180.0 -DE/DX = 0.0 ! ! D4 D(2,1,6,4) -120.0 -DE/DX = 0.0 ! ! D5 D(2,1,6,5) 120.0 -DE/DX = 0.0 ! ! D6 D(3,1,5,4) 90.0 -DE/DX = 0.0 ! ! D7 D(4,1,6,5) -120.0 -DE/DX = 0.0 !
XYZ file
6 PF<sub>5</sub> 1 P1 0.0000 0.0000 0.0000 P 2 F2 0.0000 1.5693 0.0000 F 3 F3 0.0000 0.0000 1.5970 F 4 F4 -1.3590 -0.7846 0.0000 F 5 F5 1.3590 -0.7846 0.0000 F 6 F6 0.0000 0.0000 -1.5970 F
Level 2 Lab classes: DFT calculations
BH3 optimisation data
Method: B3LYP
Basis set: 6-31G(d,p)
Calculations: OPTF
Point group:
Optimised BH3 molecule
test molecule |
Item Value Threshold Converged?
Maximum Force 0.000203 0.000450 YES
RMS Force 0.000098 0.000300 YES
Maximum Displacement 0.000710 0.001800 YES
RMS Displacement 0.000412 0.001200 YES
Predicted change in Energy=-1.365987D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1928 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1924 -DE/DX = 0.0 !
! R3 R(1,4) 1.1926 -DE/DX = -0.0002 !
! A1 A(2,1,3) 119.9864 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0146 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.999 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
NH3BH3 optimisation data
Method: B3LYP
Basis set: 6-31G(d,p)
Calculations: OPTF
Point group:
Optimised NH3BH3 molecule
test molecule |
Item Value Threshold Converged?
Maximum Force 0.000121 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000564 0.001800 YES
RMS Displacement 0.000316 0.001200 YES
Predicted change in Energy=-1.705954D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.21 -DE/DX = -0.0001 !
! R2 R(2,7) 1.21 -DE/DX = -0.0001 !
! R3 R(3,7) 1.21 -DE/DX = -0.0001 !
! R4 R(4,8) 1.0186 -DE/DX = -0.0001 !
! R5 R(5,8) 1.0186 -DE/DX = -0.0001 !
! R6 R(6,8) 1.0186 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(1,7,2) 113.8743 -DE/DX = 0.0 !
! A2 A(1,7,3) 113.8743 -DE/DX = 0.0 !
! A3 A(1,7,8) 104.5969 -DE/DX = 0.0 !
! A4 A(2,7,3) 113.8743 -DE/DX = 0.0 !
! A5 A(2,7,8) 104.5969 -DE/DX = 0.0 !
! A6 A(3,7,8) 104.5969 -DE/DX = 0.0 !
! A7 A(4,8,5) 107.8689 -DE/DX = 0.0 !
! A8 A(4,8,6) 107.8689 -DE/DX = 0.0 !
! A9 A(4,8,7) 111.0296 -DE/DX = 0.0 !
! A10 A(5,8,6) 107.8689 -DE/DX = 0.0 !
! A11 A(5,8,7) 111.0296 -DE/DX = 0.0 !
! A12 A(6,8,7) 111.0296 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 180.0 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -60.0 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.0 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 180.0 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 180.0 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -60.0 -DE/DX = 0.0 !
