CID 01180512
NH3 molecule
NH3 molecule |
The NH3 molecule is one of the most impactful compounds, due to its use as an intermediate for synthesising fertilisers in agriculture. It is made from the haber process.
N-H bond distance = 1.01798 Å
H-N-H bond angle = 105.741°
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
Energy(RB3LYP): -56.55776873 a.u., -148492.43 kJ/mol
RMS Gradient Norm: 0.00000485 a.u.
Imaginary Freq: 0
Dipole Moment: 1.8466 Debye
Point Group: C3V
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986266D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Data from calculations are found here
Vibrational Modes
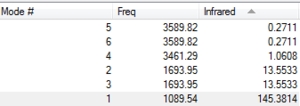
A1: NH3 has 3N - 6 modes of vibration since it is a non-linear molecule. With 4 atoms, we expected 12 - 6 = 6 modes of vibration. This is reflected by precited vibrations of the GaussView Software. A2: Modes 2 and 3 are degenerate, Modes 5 and 6 are degenerate as reflected by their frequency. A3: Mode 4 is symmetric stretch, Modes 5, 6 are asymmetric stretches. A4: Mode 4 is highly symmetric. A5: Mode 1 is umbrella mode.
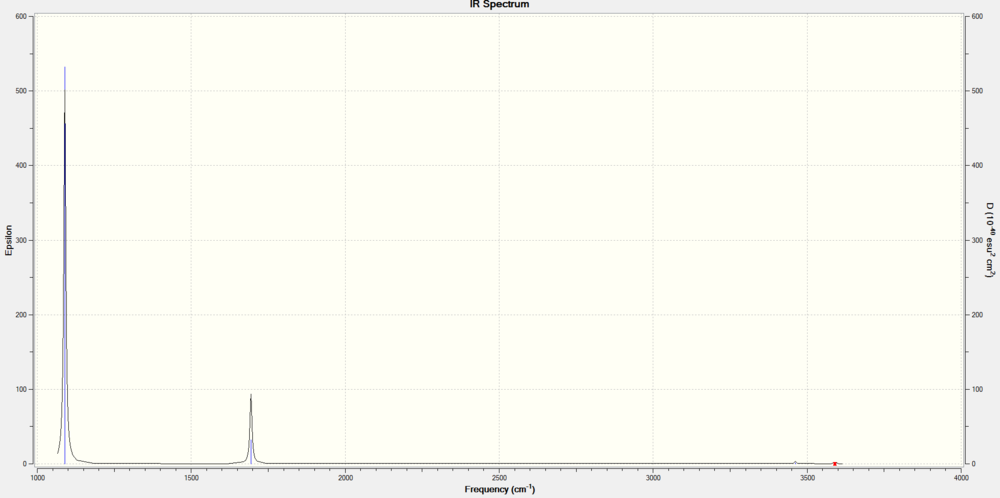
2 peaks are expected to be observed. There are 2 degenerate pairs (2,3) and (5,6) of vibrational motion out of 6 modes of vibration. In addition, the symmetric(4) and asymmetric stretches(5,6) have minimal to no change in dipole moment, resulting in extremely low intensities of their predicted IR peaks. They are predicted to be IR inactive. The vibrational modes 1 and a degenerate pair (2,3) result in a significant net change in dipole moment and can be said to be IR active. The frequencies of the IR spectrum are found to correspond to that of literature[2], but not completely, since rotational P, Q bands are observed in experimental IR spectrum, along with an additional peak. Intramolecular interactions like hydrogen bonding between NH3 is also not predicted by this software.
Charge distribution on N = - 1.125 Charge distribution on H = + 0.375
N, being more electronegative than H, would withdraw electron density to itself and have a partial negative charge, causing Hydrogen to have a net positive charge. N has a higher magnitude of charge since there is only one N for every 3 H.
H2 molecule
H2 molecule |
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
Energy(RB3LYP): -1.17853936 a.u., -3094.25 kJ/ mol
RMS Gradient Norm: 0.00000017 a.u.
Imaginary Freq: 0
Dipole Moment: 0.0000 Debye
Point Group: D∞h
Bond Length: 0.74279 Å
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Vibrational Modes
Only symmetric stretch is present in H2 molecule in accordance with 3N - 5 rule for linear molecules. No net change of dipole moment occurs, and H2 is not IR active.
N2 molecule
N2 molecule |
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
Energy(RB3LYP): -109.524129 a.u., -287555.62 kJ/mol
RMS Gradient Norm: 0.00000060 a.u.
Imaginary Freq: 0
Dipole Moment: 0.0000 Debye
Point Group: D∞h
Bond Length: 1.10550 Å
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400929D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0
Vibrational Modes
Only symmetric stretch is present in N2 molecule in accordance with 3N - 5 rule for linear molecules. No net change of dipole moment occurs, and N2 is not IR active.
Thermodynamics of Haber Process
E(NH3)= -148492.43 kJ/mol
2*E(NH3)= -296984.86 kJ/mol
E(N2)= -287555.62 kJ/mol
E(H2)= -3094.25 kJ/mol
3*E(H2)= -9282.75 kJ/mol
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.49kJ/mol
The reaction is exothermic, implying that the products have a lower energy than the reactants. N2 has the lowest energy, making it the most stable compound among the reactants and products. However, this calculated value is noticeably deviated from the literature value. [1]
- ↑ Vanderzee, C.; King, D. The Enthalpies Of Solution And Formation Of Ammonia. The Journal of Chemical Thermodynamics 1972, 4, 675-683..
of enthalpy change of formation for ammonia.
CH3OH
NH3 molecule |
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
Energy(RB3LYP): -115.7239644 a.u., -303833.29 kJ/mol
RMS Gradient Norm: 0.00000060 a.u.
Imaginary Freq: 0
Dipole Moment: 0.0000 Debye
Point Group: N.A.
Bond Length: O-H 0.96520 Å, C-H 1.1055 Å, C-O 1.41811 Å
Item Value Threshold Converged?
Maximum Force 0.000038 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.000307 0.001800 YES
RMS Displacement 0.000147 0.001200 YES
Predicted change in Energy=-1.416134D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.093 -DE/DX = 0.0 !
! R2 R(1,3) 1.1006 -DE/DX = 0.0 !
! R3 R(1,4) 1.1006 -DE/DX = 0.0 !
! R4 R(1,5) 1.4181 -DE/DX = 0.0 !
! R5 R(5,6) 0.9652 -DE/DX = 0.0 !
! A1 A(2,1,3) 107.8995 -DE/DX = 0.0 !
! A2 A(2,1,4) 107.8995 -DE/DX = 0.0 !
! A3 A(2,1,5) 106.9041 -DE/DX = 0.0 !
! A4 A(3,1,4) 108.2583 -DE/DX = 0.0 !
! A5 A(3,1,5) 112.8278 -DE/DX = 0.0 !
! A6 A(4,1,5) 112.8278 -DE/DX = 0.0 !
! A7 A(1,5,6) 107.7374 -DE/DX = 0.0 !
! D1 D(2,1,5,6) 179.9999 -DE/DX = 0.0 !
! D2 D(3,1,5,6) 61.5463 -DE/DX = 0.0 !
! D3 D(4,1,5,6) -61.5465 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Data from calculations are found here
Vibrational Modes
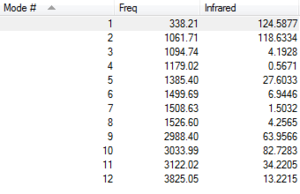
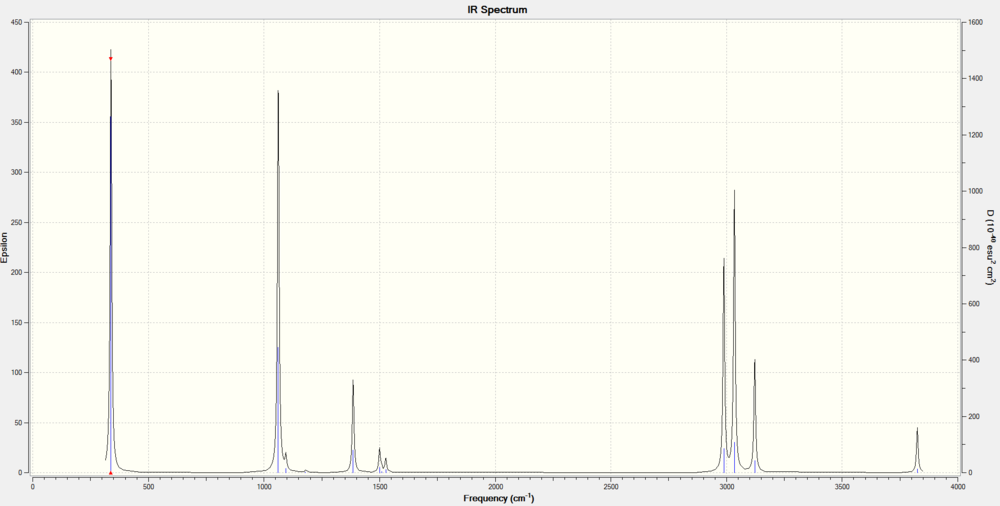
Charge Distribution
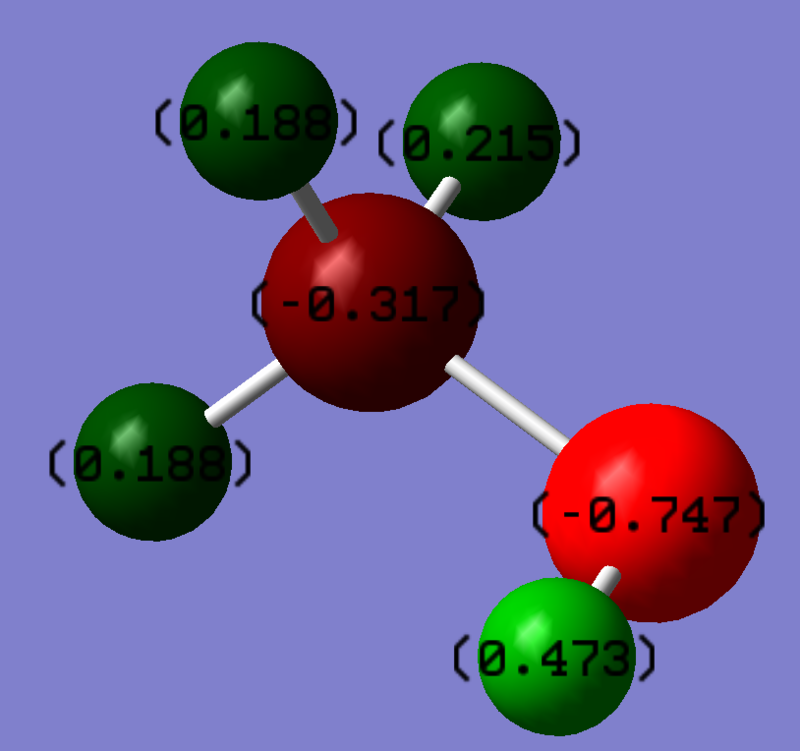
The oxygen atom is more electronegative than carbon and hydrogen, making carbon electron deficient, and hydrogen acidic.
Molecular Orbitals

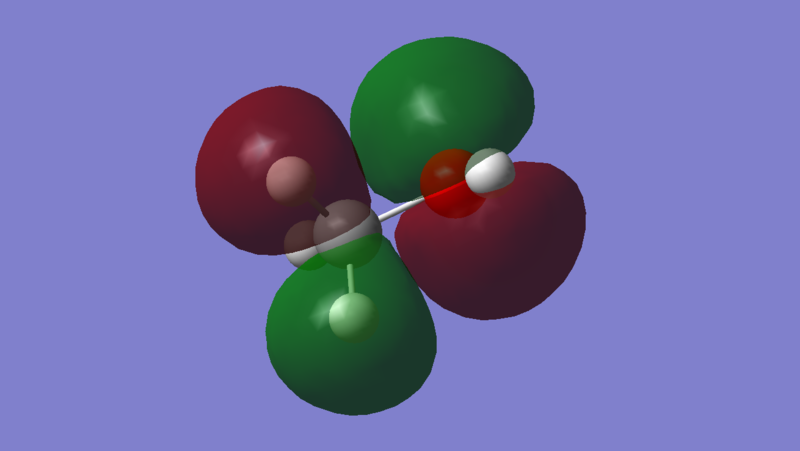
The LUMO of methanol is MO 10, and it is a sigma anti-bonding orbital predominantly between 3S orbital on Oxygen and 1S orbital on Hydrogen. As observed from the Molecular Orbital Coefficients(MO diagram too clustered), the orbital contribution from Hydrogen is greater than the orbital contribution from Oxygen. This implies that the electrophilic site lies on the Hydrogen since a base or a nucleophile will penetrate the LUMO sigma anti-bonding orbital to abstract a Hydrogen from methanol. This is expected since Oxygen is more electronegative than Hydrogen, making methanol slightly acidic.
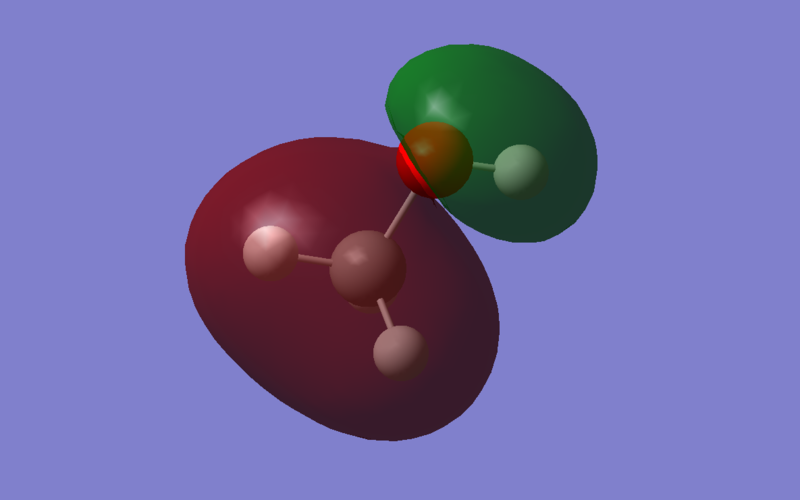
The HOMO of methanol is MO 9, and it is an anti-bonding orbital predominantly between 2Pz orbital carbon and 2Pz orbital oxygen. As observed by using the Molecular Orbital Coefficients, the orbital contribution from Oxygen is greater than the orbital contribution from Carbon. This implies that the nucleophilic site lies on the oxygen, which is expected since oxygen is more electronegative than carbon. The p orbital of carbon is out of phase relative to the p orbital of oxygen.
The S and P orbitals of Carbon overlap in phase with the 1s orbitals of Hydrogen, and the 2s orbitals of Oxygen overlap with the 1s orbitals of the alcoholic Hydrogen. However, The carbon orbitals and oxygen orbitals are out of phase. This is an anti-bonding orbital.

The Pz orbitals of Carbon side on overlap with Pz orbitals of Oxygen, resulting in π bonding. The orbital coefficients of carbon are higher than the orbital coefficients of Oxygen
Molecular Orbital Coefficients:
1 2 3 4 5
O O O O O
Eigenvalues -- -19.14032 -10.22504 -1.01058 -0.67799 -0.50124
1 1 C 1S 0.00001 0.99292 -0.07445 -0.18046 0.02598
2 2S 0.00023 0.04881 0.14153 0.36653 -0.04959
3 2PX -0.00034 -0.00055 -0.10464 0.06398 -0.23158
4 2PY 0.00011 0.00001 0.00426 0.01628 0.21365
5 2PZ 0.00000 0.00000 0.00000 0.00000 0.00000
6 3S -0.00156 -0.01345 0.04856 0.31820 -0.05786
7 3PX 0.00079 -0.00047 0.00346 0.02931 -0.07910
8 3PY -0.00051 -0.00006 -0.00865 -0.00366 0.09334
9 3PZ 0.00000 0.00000 0.00000 0.00000 0.00000
10 4XX 0.00029 -0.00884 0.01887 -0.01412 0.02177
11 4YY 0.00005 -0.00900 -0.01088 -0.00590 -0.00188
12 4ZZ 0.00007 -0.00902 -0.01190 -0.00780 -0.01433
13 4XY -0.00005 0.00000 -0.00049 -0.00238 -0.00986
14 4XZ 0.00000 0.00000 0.00000 0.00000 0.00000
15 4YZ 0.00000 0.00000 0.00000 0.00000 0.00000
16 2 H 1S 0.00011 -0.00018 0.02690 0.14486 0.02753
17 2S 0.00021 0.00262 0.00883 0.05104 0.01578
18 3PX 0.00006 0.00007 -0.00286 -0.00360 -0.00412
19 3PY 0.00003 0.00009 -0.00375 -0.01181 0.00185
20 3PZ 0.00000 0.00000 0.00000 0.00000 0.00000
21 3 H 1S 0.00002 -0.00018 0.02575 0.13451 -0.11941
22 2S -0.00006 0.00256 0.00226 0.04373 -0.07691
23 3PX 0.00003 0.00005 -0.00265 -0.00317 -0.00066
24 3PY 0.00000 -0.00005 0.00233 0.00599 0.00006
25 3PZ -0.00005 -0.00009 0.00336 0.00986 -0.00584
26 4 H 1S 0.00002 -0.00018 0.02575 0.13451 -0.11941
27 2S -0.00006 0.00256 0.00226 0.04373 -0.07691
28 3PX 0.00003 0.00005 -0.00265 -0.00317 -0.00066
29 3PY 0.00000 -0.00005 0.00233 0.00599 0.00006
30 3PZ 0.00005 0.00009 -0.00336 -0.00986 0.00584
31 5 O 1S 0.99282 -0.00010 -0.20340 0.06005 -0.03947
32 2S 0.02609 -0.00012 0.44920 -0.13740 0.09216
33 2PX 0.00042 -0.00011 0.06225 0.18356 0.28342
34 2PY -0.00104 0.00002 -0.09457 0.09398 0.31779
35 2PZ 0.00000 0.00000 0.00000 0.00000 0.00000
36 3S 0.01108 0.00159 0.42661 -0.16859 0.14669
37 3PX 0.00022 0.00004 0.03963 0.08296 0.12761
38 3PY -0.00004 0.00027 -0.03301 0.03731 0.16323
39 3PZ 0.00000 0.00000 0.00000 0.00000 0.00000
40 4XX -0.00796 -0.00076 0.00842 0.01661 0.01208
41 4YY -0.00772 -0.00017 -0.00030 -0.00708 -0.02116
42 4ZZ -0.00787 0.00002 -0.00997 0.00059 0.00150
43 4XY 0.00010 0.00006 -0.00243 -0.00672 0.00010
44 4XZ 0.00000 0.00000 0.00000 0.00000 0.00000
45 4YZ 0.00000 0.00000 0.00000 0.00000 0.00000
46 6 H 1S 0.00022 0.00011 0.14306 -0.12500 -0.21191
47 2S -0.00122 0.00047 0.01627 -0.04635 -0.13753
48 3PX -0.00015 0.00021 0.01119 -0.00215 -0.00194
49 3PY -0.00023 0.00006 0.02022 -0.01306 -0.01180
50 3PZ 0.00000 0.00000 0.00000 0.00000 0.00000
6 7 8 9 10
O O O O V
Eigenvalues -- -0.43327 -0.41891 -0.32942 -0.26493 0.07706
1 1 C 1S 0.00000 -0.00543 0.01363 0.00000 0.03387
2 2S 0.00000 0.01119 -0.03358 0.00000 -0.03544
3 2PX 0.00000 -0.27329 0.11695 0.00000 -0.19984
4 2PY 0.00000 -0.30662 -0.26991 0.00000 0.05156
5 2PZ 0.42016 0.00000 0.00000 -0.17875 0.00000
6 3S 0.00000 0.02231 -0.03258 0.00000 -0.64335
7 3PX 0.00000 -0.11211 0.03513 0.00000 -0.57069
8 3PY 0.00000 -0.15400 -0.07932 0.00000 0.07905
9 3PZ 0.17323 0.00000 0.00000 -0.02245 0.00000
10 4XX 0.00000 0.01845 -0.00832 0.00000 -0.00787
11 4YY 0.00000 -0.02006 -0.00488 0.00000 0.01773
12 4ZZ 0.00000 0.00052 0.01518 0.00000 0.01069
13 4XY 0.00000 -0.00836 -0.02462 0.00000 -0.00338
14 4XZ 0.00249 0.00000 0.00000 -0.03162 0.00000
15 4YZ -0.01587 0.00000 0.00000 0.01328 0.00000
16 2 H 1S 0.00000 -0.23670 -0.17712 0.00000 0.00750
17 2S 0.00000 -0.18682 -0.18524 0.00000 0.31414
18 3PX 0.00000 0.00113 0.00336 0.00000 -0.00732
19 3PY 0.00000 0.00617 0.00333 0.00000 0.00477
20 3PZ 0.00779 0.00000 0.00000 -0.00486 0.00000
21 3 H 1S -0.20307 0.03695 0.11390 0.14857 0.00824
22 2S -0.17208 0.02411 0.13837 0.19828 0.46972
23 3PX 0.00461 -0.00480 0.00065 0.00034 -0.00597
24 3PY -0.00539 -0.00425 -0.00360 0.00311 -0.00351
25 3PZ -0.00227 0.00174 0.00533 0.00115 -0.00306
26 4 H 1S 0.20307 0.03695 0.11390 -0.14857 0.00824
27 2S 0.17208 0.02411 0.13837 -0.19829 0.46972
28 3PX -0.00461 -0.00480 0.00065 -0.00034 -0.00597
29 3PY 0.00539 -0.00425 -0.00360 -0.00311 -0.00351
30 3PZ -0.00227 -0.00174 -0.00533 0.00115 0.00306
31 5 O 1S 0.00000 0.03131 -0.06665 0.00000 0.09013
32 2S 0.00000 -0.05803 0.12995 0.00000 -0.11259
33 2PX 0.00000 0.34112 -0.19977 0.00000 0.01093
34 2PY 0.00000 -0.12929 0.42082 0.00000 0.25062
35 2PZ 0.25642 0.00000 0.00000 0.59487 0.00000
36 3S 0.00000 -0.14065 0.32712 0.00000 -1.11893
37 3PX 0.00000 0.19340 -0.14321 0.00000 0.00348
38 3PY 0.00000 -0.08315 0.28144 0.00000 0.44926
39 3PZ 0.16208 0.00000 0.00000 0.45629 0.00000
40 4XX 0.00000 0.00966 -0.00899 0.00000 0.03673
41 4YY 0.00000 0.00558 -0.03088 0.00000 0.02681
42 4ZZ 0.00000 0.00106 -0.00012 0.00000 0.04788
43 4XY 0.00000 -0.02405 0.00764 0.00000 0.00601
44 4XZ 0.01420 0.00000 0.00000 0.00830 0.00000
45 4YZ -0.01107 0.00000 0.00000 -0.02046 0.00000
46 6 H 1S 0.00000 -0.04499 -0.15221 0.00000 0.11132
47 2S 0.00000 -0.04683 -0.12187 0.00000 1.21515
48 3PX 0.00000 0.01073 -0.01014 0.00000 -0.00085
49 3PY 0.00000 -0.00761 0.00212 0.00000 -0.00669
50 3PZ 0.00855 0.00000 0.00000 0.01974 0.00000
11 12 13 14
V V V V
Eigenvalues -- 0.13104 0.16702 0.17527 0.20619
1 1 C 1S -0.15487 -0.01282 0.00000 0.05890
2 2S 0.21297 0.01079 0.00000 -0.08870
3 2PX 0.06530 -0.12508 0.00000 0.29847
4 2PY -0.01197 -0.40470 0.00000 -0.12891
5 2PZ 0.00000 0.00000 -0.45775 0.00000
6 3S 2.39420 0.09431 0.00000 -0.69821
7 3PX 0.17130 -0.47543 0.00000 1.25510
8 3PY -0.01026 -1.21554 0.00000 -0.58663
9 3PZ 0.00000 0.00000 -1.30257 0.00000
10 4XX -0.01850 -0.01083 0.00000 0.02647
11 4YY -0.00605 0.01975 0.00000 -0.00860
12 4ZZ -0.01207 -0.00922 0.00000 -0.02545
13 4XY -0.00168 0.01142 0.00000 -0.00791
14 4XZ 0.00000 0.00000 0.00201 0.00000
15 4YZ 0.00000 0.00000 -0.01613 0.00000
16 2 H 1S -0.02291 0.11371 0.00000 -0.01780
17 2S -1.05297 1.74935 0.00000 0.48445
18 3PX -0.00364 -0.00071 0.00000 0.00975
19 3PY -0.00326 0.00510 0.00000 -0.00405
20 3PZ 0.00000 0.00000 -0.01324 0.00000
21 3 H 1S -0.02672 -0.04197 -0.07143 -0.06163
22 2S -1.08974 -0.71549 -1.50910 -0.43562
23 3PX -0.00351 -0.00614 -0.00214 0.01361
24 3PY 0.00076 -0.00813 0.00809 -0.00276
25 3PZ 0.00424 0.00485 -0.00011 0.00768
26 4 H 1S -0.02672 -0.04197 0.07143 -0.06163
27 2S -1.08974 -0.71549 1.50911 -0.43562
28 3PX -0.00351 -0.00614 0.00214 0.01361
29 3PY 0.00076 -0.00813 -0.00809 -0.00276
30 3PZ -0.00424 -0.00485 -0.00011 -0.00768
31 5 O 1S 0.05177 0.00938 0.00000 -0.04560
32 2S -0.04318 -0.01849 0.00000 0.06015
33 2PX -0.08786 -0.14206 0.00000 0.37998
34 2PY 0.12857 0.03320 0.00000 0.14648
35 2PZ 0.00000 0.00000 0.10483 0.00000
36 3S -0.71684 -0.13485 0.00000 0.62753
37 3PX -0.20959 -0.23816 0.00000 0.83424
38 3PY 0.21491 0.11718 0.00000 0.28942
39 3PZ 0.00000 0.00000 0.23356 0.00000
40 4XX 0.03557 -0.00436 0.00000 -0.00182
41 4YY 0.02264 0.00249 0.00000 -0.02219
42 4ZZ 0.02819 -0.00080 0.00000 -0.02095
43 4XY 0.00646 -0.01091 0.00000 -0.01221
44 4XZ 0.00000 0.00000 -0.01083 0.00000
45 4YZ 0.00000 0.00000 -0.01209 0.00000
46 6 H 1S 0.05604 -0.04447 0.00000 0.06133
47 2S 0.51277 -0.28369 0.00000 0.88765
48 3PX -0.00771 0.00635 0.00000 0.00765
49 3PY -0.00467 0.00082 0.00000 -0.00126
50 3PZ 0.00000 0.00000 0.00708 0.00000
