At3815
In Introduction to Molecular Modelling 2, the use of GaussView 5.0 is used to obtain information and establish optimised structures of various species, and to investigate the transitions states and intermediates that cannot be observed by a chemical reaction. In this lab, an investigation of the NH3 Molecule was carried out and the following data was obtained.
NH3 Molecule
Optimisation of molecule
| NH3 Molecule | ||||
|---|---|---|---|---|
| ||||
| More Info | ||||
| Calculation Method | RB3LYP | |||
| Basis Set | 6-31g(d,p) | |||
| Final Energy | -56.55776873 a.u. | |||
| RMS Gradient | 0.00000485 a.u. | |||
| Point Group | C3v | |||
Bond Length (N-H): 1.01798Å
Bond Angle (H-N-H): 105.741°
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986298D-10 Optimization completed.
The optimisation file is linked to here.
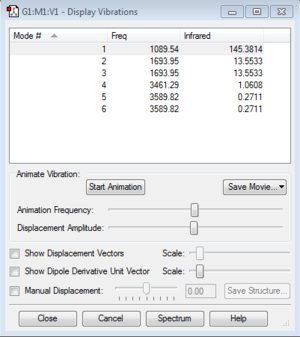
Display Vibrations
Charge Analysis
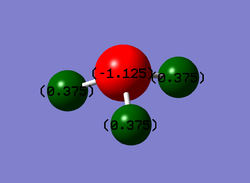
Due to a higher electronegativity in nitrogen atom compared to hydrogen atom, a negative charge is expected for the N atom and a positive charge for the H atom.
Reaction Energies
H2 Molecule
Optimisation of Molecule
| H2 Molecule | ||||
|---|---|---|---|---|
| ||||
| More Info | ||||
| Calculation Method | RB3LYP | |||
| Basis Set | 6-31g(d,p) | |||
| Final Energy | -1.17853936 a.u. | |||
| RMS Gradient | 0.00000017 a.u. | |||
| Point Group | D∞h | |||
Bond Length (H-H): 0.74279Å
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13 Optimization completed.
The optimisation file is linked to here.
Display Vibrations
N2 Molecule
Optimisation of Molecule
| N2 Molecule | ||||
|---|---|---|---|---|
| ||||
| More Info | ||||
| Calculation Method | RB3LYP | |||
| Basis Set | 6-31g(d,p) | |||
| Final Energy | -109.52412868 a.u. | |||
| RMS Gradient | 0.00000060 a.u. | |||
| Point Group | D∞h | |||
Bond Length (N-N): 1.10550Å
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401111D-13 Optimization completed.
The optimisation file is linked to here.
Display Vibrations

Haber-Bosch process
The energy for the reaction of N2 + 3H2 -> 2NH3 can be calculated using the below energies.
- E(NH3)= -56.55776873 a.u.
- 2*E(NH3)= -113.11553746 a.u.
- E(N2)= -109.52412868 a.u.
- E(H2)= -1.17853936 a.u.
- 3*E(H2)= -3.53561808 a.u.
- ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579070 a.u.= -146.47 kJ/mol
Since ΔE is negative which shows that the forward reaction is exothermic, the ammonia product is more stable than the gaseous reactants.
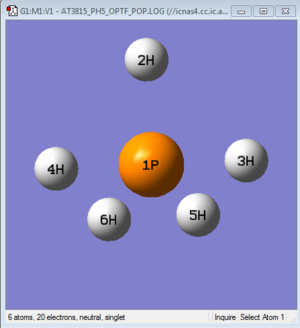
PH5 Molecule
The small molecule I have chosen for investigation is phosphorane, PH5 which adopts a trigonal bipyramidal molecular geometry. A similar type of analysis using GaussView 5.0 was carried out for PH5 as with NH3 and the following information was obtained.
Optimisation of Molecule
| PH5 Molecule | ||||
|---|---|---|---|---|
| ||||
| More Info | ||||
| Calculation Method | RB3LYP | |||
| Basis Set | 6-31g(d,p) | |||
| Final Energy | -344.25491049a.u. | |||
| RMS Gradient | 0.00000471 a.u. | |||
| Point Group | D3h | |||
The optimisation file is linked to here.
Bond Lengths (P-2H, P-5H, P-6H): 1.43316Å ; (P-3H, P-4H): 1.48687Å
Bond Angles (2H-P-3H, 2H-P-4H, 3H-P-6H, 4H-P-5H) = 90° ; (5H-P-6H) = 120°
Display Vibrations
It can be seen that PH5 has 12 vibration modes. However, there are only 8 distinct frequencies. There are 4 pairs of degenerate modes: 1 & 2, 4 & 5, 6 & 7 and 11 & 12. In the IR spectrum, only 4 distinct bands can be observed as the intensities for mode 1, 2, 6, 7, 8 and 10 are zero or too low to be observed. Modes 1 to 7 are "bending" vibrations while modes 8 to 12 are "bond stretch" vibrations.
Charge Analysis
As phosphorous is less electronegative than hydrogen, the phosphorous atom is expected to carry a positive charge while the neighbouring hydrogen atoms carry a negative charge. Since PH5 is a neutral molecule, the sum of the partial negative charges on all the hydrogens are similar to the partical positive charge on phosphorous.
Molecular Orbitals
Further Analysis of PH5 Molecule
Through running the structure of PH3 through GaussView 5.0, the following information was gathered about the molecule.
| PH3 Molecule | ||||
|---|---|---|---|---|
| ||||
| More Info | ||||
| Calculation Method | RB3LYP | |||
| Basis Set | 6-31g(d,p) | |||
| Final Energy | -343.14506758a.u. | |||
| RMS Gradient | 0.00009144 a.u. | |||
| Point Group | C3v | |||
The geometry of PH3 is similar to NH3 with a pyramidal shape.
From the information of PH3 gathered, the energy of the formation of PH5 through the reaction of PH3 + H2 -> PH5 can be calculated using the following energies.
- E(PH5)= -344.25491049a.u.
- E(H2)= -1.17853936 a.u.
- E(PH3)=-343.14506758a.u.
- ΔE=E(PH5)-[E(PH3)+E(H2)]= 0.06869645 a.u.
As ΔE is positive, this shows that the formation of PH5 is endothermic and that the product is of a higher energy than the reactants, thus PH5 is not a stable molecule and generally does not exist.
On the other hand, other bipyramidal phosphorane molecules such as PF5 is a more stable molecule with a much lower energy of -840.67634601 a.u.