01409469
Introduction
Gaussview was utilised in the optimisation of molecules such as NH3, N2 and ClF3. This Wikipedia page documents the data obtained from Gaussview.
NH3
SUMMARY
| Parameter | Result |
|---|---|
| Molecule name | Ammonia |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -56.55776873 a.u. |
| RMS gradient norm | 0.00000485 a.u. |
| Point group | C3V |
| Dipole moment | 1.8467 Debye |
ITEM TABLE
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986267D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Ammonia molecule |
OPTIMISATION
| Optimised parameter | Result |
|---|---|
| N-H bond distance | 1.01798Å |
| H-N-H bond angle | 105.741° |
| Optimisation/frequency file | Media:RCHAN_NH3_OPTF_POP.LOG |
VIBRATIONS
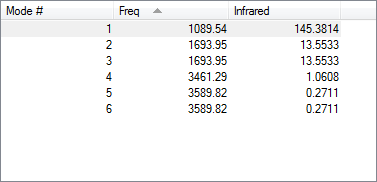
| Parameter | Result |
|---|---|
| Degenerate modes | Modes 2 and 3 Modes 5 and 6 |
| Bending vibrations | 1,2,3 |
| Bond stretch vibrations | 4,5,6 |
| Highly symmetric mode | 4 |
| Umbrella mode | 1 |
Following the 3N-6 rule, the expected number of modes is 6.
Expected number of bands in an experimental spectrum of gaseous ammonia: 2
The expected number of bands for ammonia is 2 because only modes 1, 2 and 3 have a change in dipole moment and are IR active. Modes 2 and 3 are degenerate and correspond to the same peak. As a result only 2 peaks will be seen in the IR spectrum.
ATOMIC CHARGES
| Atom | Charge |
|---|---|
| Nitrogen | -1.125 |
| Hydrogen | 0.375 |
A negative charge is expected for N and a positive charge is expected for H. On the Pauling scale for electronegativity, N has an electronegativity of 3 while H has a electronegativity of 2.1. N is more electronegative than H and is expected to have a negative charge while the more electropositive H has a positive charge.
N2
SUMMARY
| Parameter | Result |
|---|---|
| Molecule name | Nitrogen |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -109.52412868 a.u. |
| RMS gradient norm | 0.00000060 a.u. |
| Point group | D∞H |
| Dipole moment | 0.0000 Debye |
ITEM TABLE
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401071D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Nitrogen molecule |
OPTIMISATION
| Optimised parameter | Result |
|---|---|
| N-N bond distance | 1.10550 Å |
| Optimisation/frequency file | Media:RCHAN_N2_OPTF.LOG |
VIBRATIONS
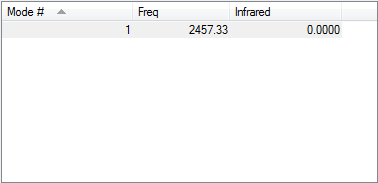
Following the 3N-5 rule, the expected number of vibrational modes is 1.
This vibration is a symmetric bond stretch.
H2
SUMMARY
| Parameter | Result |
|---|---|
| Molecule name | Hydrogen |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -1.17853936 a.u. |
| RMS gradient norm | 0.00000017 a.u. |
| Point group | D∞H |
| Dipole moment | 0.0000 Debye |
ITEM TABLE
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Hydrogen molecule |
OPTIMISATION
| Optimised parameter | Result |
|---|---|
| H-H bond distance | 0.74279 Å |
| Optimisation/frequency file | Media:RCHAN_H2_OPTF.LOG |
VIBRATIONS
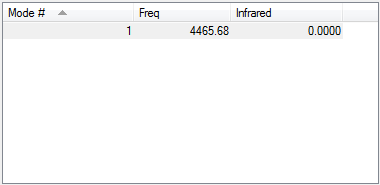
Following the 3N-5 rule, the expected number of vibrational modes is 1.
This vibration is a symmetric stretch.
Haber-Bosch Reaction Energy Calculation
| Calculation | Calculated values |
|---|---|
| E(NH3) | -56.55776873 a.u. |
| 2*E(NH3) | -113.1155375 a.u. |
| E(N2) | -109.52412868 a.u. |
| E(H2) | -1.17853936 a.u. |
| 3*E(H2) | -3.53561808 a.u. |
| ΔE=2*E(NH3)-[E(N2)+3*E(H2)] | -0.0557907 a.u. = -146.48 kJ/mol |
The ammonia product is more stable than the gaseous reactants because the ammonia product is lower in energy than the reactants.
Independence: ClF3
SUMMARY
| Parameter | Result |
|---|---|
| Molecule name | Chlorine trifluoride |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -759.46531688 a.u. |
| RMS gradient norm | 0.00002465 a.u. |
| Point group | C2V |
| Dipole moment | 0.8386 Debye |
ITEM TABLE
Item Value Threshold Converged?
Maximum Force 0.000050 0.000450 YES
RMS Force 0.000028 0.000300 YES
Maximum Displacement 0.000204 0.001800 YES
RMS Displacement 0.000134 0.001200 YES
Predicted change in Energy=-1.250255D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.7286 -DE/DX = 0.0 !
! R2 R(1,3) 1.6514 -DE/DX = 0.0 !
! R3 R(1,4) 1.7286 -DE/DX = 0.0 !
! A1 A(2,1,3) 87.1404 -DE/DX = 0.0 !
! A2 A(3,1,4) 87.1404 -DE/DX = 0.0 !
! A3 L(2,1,4,3,-1) 174.2807 -DE/DX = 0.0 !
! A4 L(2,1,4,3,-2) 180.0 -DE/DX = 0.0 !
! D1 D(2,1,3,4) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
chlorine trifluoride molecule |
OPTIMISATION
| Optimised parameter | Result |
|---|---|
| Cl-F bond distance | 1.72863 Å 1.65143 Å |
| F-Cl-F bond angle | 87.140 ° |
| Optimisation/frequency file | Media: RCHAN_CLF3_OPTF.LOG |
VIBRATIONS
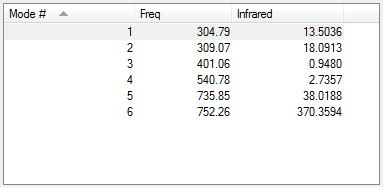
| Parameter | Result |
|---|---|
| Bending vibrations | 1,2,3 |
| Bond stretch vibrations | 4,5,6 |
Following the 3N-6 rule, the expected number of modes is 6.
None of the vibrations are degenerate.
ATOMIC CHARGES
| Atom | Charge |
|---|---|
| Fluorine | -0.316 -0.454 |
| Chlorine | +1.225 |
A negative charge is expected for F and a positive charge is expected for Cl. On the Pauling scale for electronegativity, Cl has an electronegativity of 3.16 while F has a electronegativity of 3.98. F is more electronegative than Cl and is expected to have a negative charge while the more electropositive Cl has a positive charge.
There are two different values for the charges on fluorine. This is because the fluorines are in two different environments based on whether they are in the axial position or equatorial position.
MOLECULAR ORBITALS
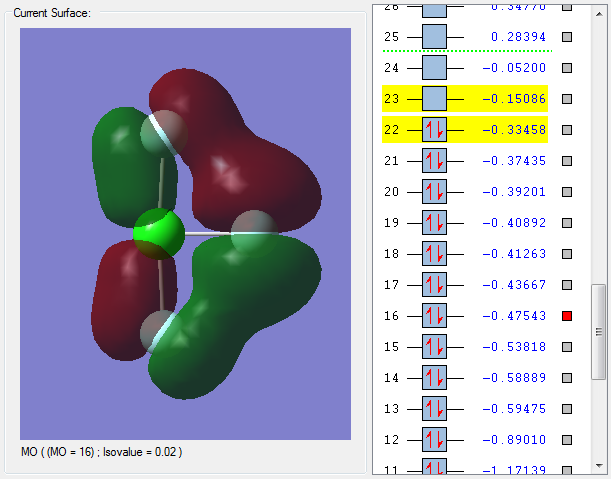
For the axial Cl-F bond, the orbital from Cl and the orbital of F have combined in phase to give a bonding molecular orbital. The bonding molecular orbital has a positive effect on bonding and strengthens the axial Cl-F bond.
For the equatorial Cl-F bond, the orbital from Cl and the orbital from F have combined out of phase resulting in an antibonding orbital. The presence of a node at the Cl-F bond indicates an antibonding molecular orbital. The antibonding molecular orbital has a negative effect on bonding and weakens the equatorial Cl-F bond.
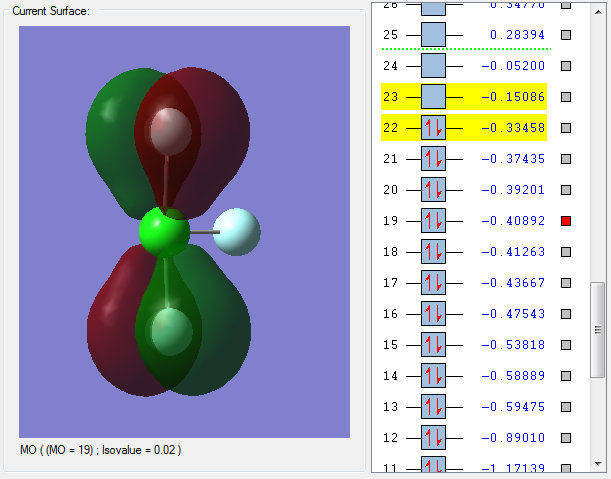
The orbital from Cl and the orbital of F have combined in phase to give a bonding molecular orbital. The presence of a node on the Cl atom rather than the Cl-F bond indicates that the Cl orbital and F orbital have combined in phase to give a bonding molecular orbital. The bonding molecular orbital has a positive effect on bonding and strengthens the axial Cl-F bond.
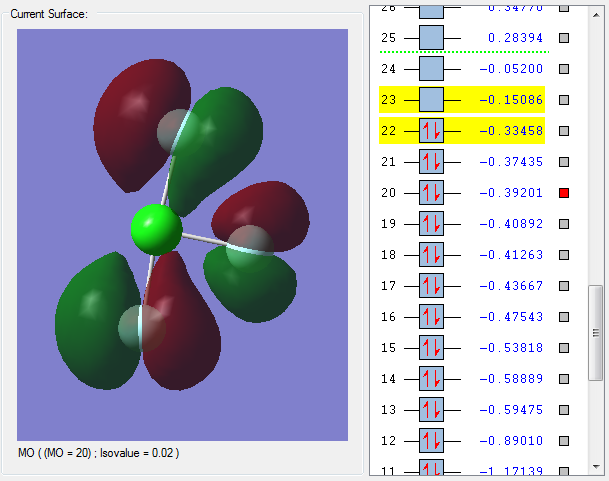
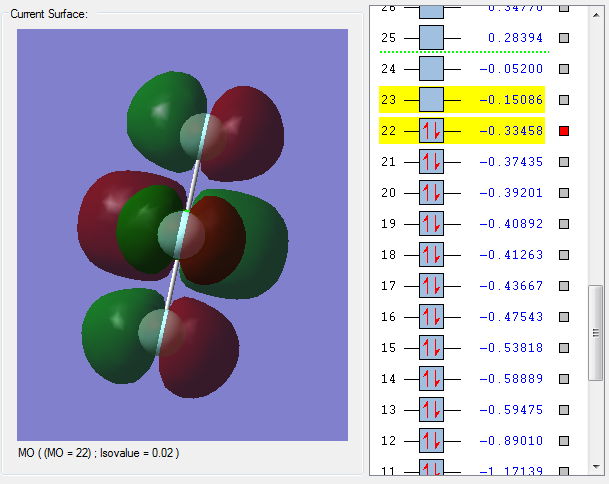
MO 22 is the highest occupied molecular orbital (HOMO). The HOMO is a bonding molecular orbital as the nodes are on the F atoms rather than in between the Cl-F bonds. Bonding molecular orbitals have a positive effect on bonding and strengthen the Cl-F bond.
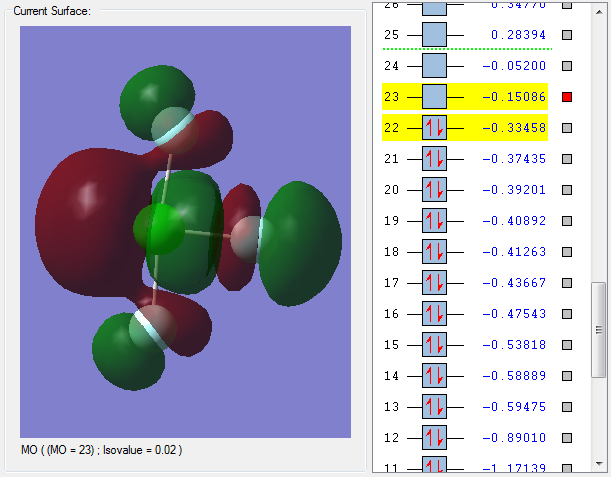
MO 23 is the lowest unoccupied molecular orbital (LUMO). This is an antibonding molecular obrital as the orbitals on Cl and the orbitals on F have combined out of phase. This is evident by the presence of nodes on the Cl-F bond. Antibonding molecular orbitals have a negative effect on bonding and weaken the Cl-F bond.
Choice of small molecule: F2
SUMMARY
| Parameter | Result |
|---|---|
| Molecule name | Fluorine |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -199.49825218 a.u. |
| RMS gradient norm | 0.00007365 a.u. |
| Point group | D∞H |
| Dipole moment | 0.0000 Debye |
ITEM TABLE
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995024D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4028 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Fluorine molecule |
OPTIMISATION
| Optimised parameter | Result |
|---|---|
| F-F bond distance | 1.40281Å |
| Optimisation/frequency file | Media: RCHAN_F2_OPTF.LOG |
VIBRATIONS
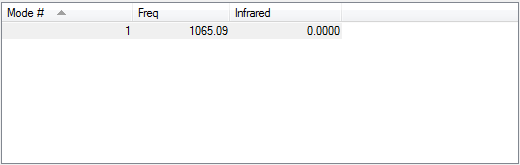
Following the 3N-5 rule, the expected number of modes is 1.
The vibration is a symmetric stretch.
ATOMIC CHARGES
Charge on F atom: 0.000
The charge on F is expected because F2 is a homonuclear diatomic molecule. As such, there is an even distribution of charge on the molecule.
MOLECULAR ORBITALS
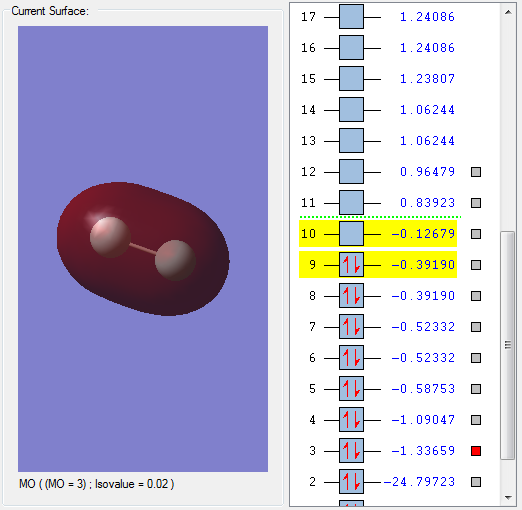
MO 3 is a bonding orbital formed by the interaction between two 2s orbitals which have combined in phase.
The MO is fully occupied with 2 electrons and deep in energy.
Because the MO is a bonding MO, this results in the formation of a bond between the 2 fluorine atoms.
MO 3 has a positive effect on bonding and strengthens the F-F bond.
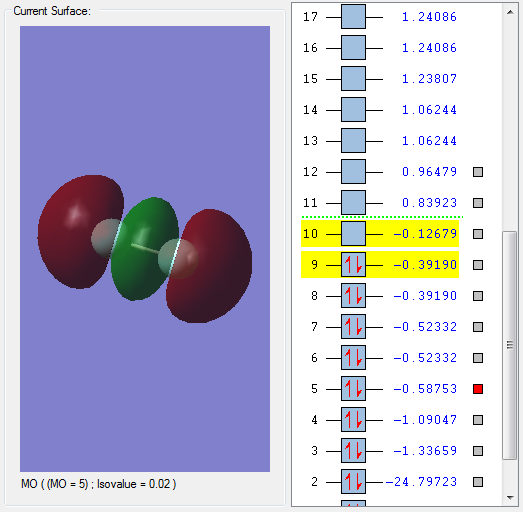
MO 5 is a bonding orbital formed by the interaction between two 2pz orbitals which have combined in phase.
The MO is fully occupied with 2 electrons and deep in energy.
Because the MO is a bonding MO, this results in the formation of a bond between the 2 fluorine atoms.
MO 5 has a positive effect on bonding and strengthens the F-F bond.
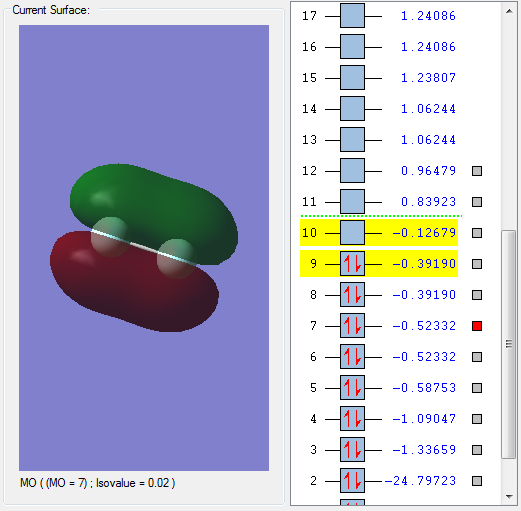
MO 7 is a bonding orbital formed by the interaction between two 2px orbitals which have combined in phase.
The MO is fully occupied with 2 electrons and deep in energy.
Because the MO is a bonding MO, this results in the formation of a bond between the 2 fluorine atoms.
MO 7 has a positive effect on bonding and strengthens the F-F bond.
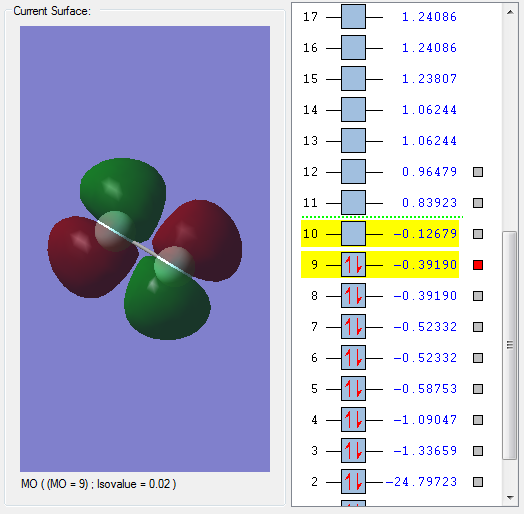
MO 9 is an antibonding orbital formed by the interaction between two 2py orbitals which have combined out-of-phase.
The MO is fully occupied with 2 electrons and is the highest occupied molecular orbital (HOMO). The MO is in the HOMO/LUMO region.
Because the MO is an antibonding MO, this results in the breakage of a bond between the 2 fluorine atoms.
MO 9 has a negative effect on bonding and weakens the F-F bond.
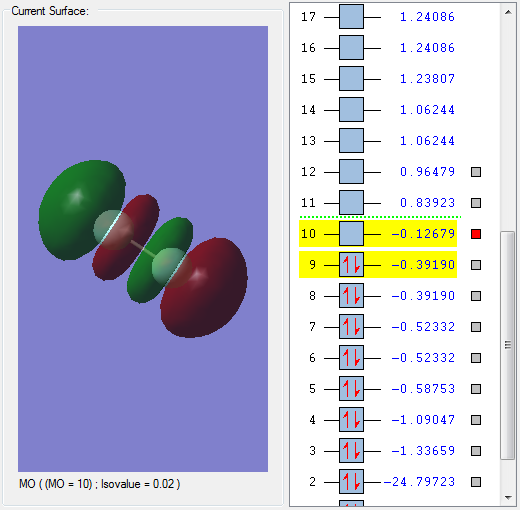
MO 10 is an antibonding orbital formed by the interaction between two 2pz orbitals which have combined out-of-phase.
The MO is unoccupied with no electrons and is the lowest unoccupied molecular orbital (LUMO). The MO is in the HOMO/LUMO region.
Because the MO is an antibonding MO, this results in the breakage of a bond between the 2 fluorine atoms.
MO 10 has a negative effect on bonding and weakens the F-F bond.
